Characterising plasmacytoid and myeloid AXL+ SIGLEC-6+ dendritic cell functions and their interactions with HIV
- PMID: 38924030
- PMCID: PMC11233022
- DOI: 10.1371/journal.ppat.1012351
Characterising plasmacytoid and myeloid AXL+ SIGLEC-6+ dendritic cell functions and their interactions with HIV
Abstract
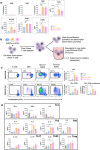
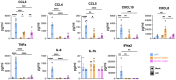
AXL+ Siglec-6+ dendritic cells (ASDC) are novel myeloid DCs which can be subdivided into CD11c+ and CD123+ expressing subsets. We showed for the first time that these two ASDC subsets are present in inflamed human anogenital tissues where HIV transmission occurs. Their presence in inflamed tissues was supported by single cell RNA analysis of public databases of such tissues including psoriasis diseased skin and colorectal cancer. Almost all previous studies have examined ASDCs as a combined population. Our data revealed that the two ASDC subsets differ markedly in their functions when compared with each other and to pDCs. Relative to their cell functions, both subsets of blood ASDCs but not pDCs expressed co-stimulatory and maturation markers which were more prevalent on CD11c+ ASDCs, thus inducing more T cell proliferation and activation than their CD123+ counterparts. There was also a significant polarisation of naïve T cells by both ASDC subsets toward Th2, Th9, Th22, Th17 and Treg but less toward a Th1 phenotype. Furthermore, we investigated the expression of chemokine receptors that facilitate ASDCs and pDCs migration from blood to inflamed tissues, their HIV binding receptors, and their interactions with HIV and CD4 T cells. For HIV infection, within 2 hours of HIV exposure, CD11c+ ASDCs showed a trend in more viral transfer to T cells than CD123+ ASDCs and pDCs for first phase transfer. However, for second phase transfer, CD123+ ASDCs showed a trend in transferring more HIV than CD11c+ ASDCs and there was no viral transfer from pDCs. As anogenital inflammation is a prerequisite for HIV transmission, strategies to inhibit ASDC recruitment into inflamed tissues and their ability to transmit HIV to CD4 T cells should be considered.
Copyright: © 2024 Warner van Dijk et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

