Single-Cell Analysis Reveals Novel Immune Perturbations in Fibrotic Hypersensitivity Pneumonitis
- PMID: 38924775
- PMCID: PMC11568434
- DOI: 10.1164/rccm.202401-0078OC
Single-Cell Analysis Reveals Novel Immune Perturbations in Fibrotic Hypersensitivity Pneumonitis
Abstract
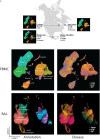
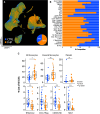
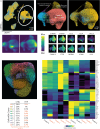
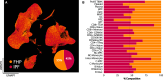
Rationale: Fibrotic hypersensitivity pneumonitis (FHP) is a debilitating interstitial lung disease driven by incompletely understood immune mechanisms. Objectives: To elucidate immune aberrations in FHP in single-cell resolution. Methods: Single-cell 5' RNA sequencing was conducted on peripheral blood mononuclear cells and BAL cells obtained from 45 patients with FHP, 63 patients with idiopathic pulmonary fibrosis (IPF), 4 patients with nonfibrotic hypersensitivity pneumonitis, and 36 healthy control subjects in the United States and Mexico. Analyses included differential gene expression (Seurat), TF (transcription factor) activity imputation (DoRothEA-VIPER), and trajectory analyses (Monocle3 and Velocyto-scVelo-CellRank). Measurements and Main Results: Overall, 501,534 peripheral blood mononuclear cells from 110 patients and control subjects and 88,336 BAL cells from 19 patients were profiled. Compared with control samples, FHP has elevated classical monocytes (adjusted-P = 2.5 × 10-3) and is enriched in CCL3hi/CCL4hi and S100Ahi classical monocytes (adjusted-P < 2.2 × 10-16). Trajectory analyses demonstrate that S100Ahi classical monocytes differentiate into SPP1hi lung macrophages associated with fibrosis. Compared with both control subjects and IPF, cells from patients with FHP are significantly enriched in GZMhi cytotoxic T cells. These cells exhibit TF activities indicative of TGFβ and TNFα and NFκB pathways. These results are publicly available at http://ildimmunecellatlas.com. Conclusions: Single-cell transcriptomics of patients with FHP uncovered novel immune perturbations, including previously undescribed increases in GZMhi cytotoxic CD4+ and CD8+ T cells-reflecting this disease's unique inflammatory T cell-driven nature-as well as increased S100Ahi and CCL3hi/CCL4hi classical monocytes also observed in IPF. Both cell populations may guide the development of new biomarkers and therapeutic interventions.
Keywords: fibrotic hypersensitivity pneumonitis; idiopathic pulmonary fibrosis; interstitial lung disease; single-cell RNA sequencing; usual interstitial pneumonia.
Figures

Comment in
-
GZMhi Cytotoxic T Cells: A Key Discovery in Fibrotic Hypersensitivity Pneumonitis.Am J Respir Crit Care Med. 2024 Nov 15;210(10):1180-1182. doi: 10.1164/rccm.202406-1194ED. Am J Respir Crit Care Med. 2024. PMID: 39078173 Free PMC article. No abstract available.
References
-
- Costabel U, Miyazaki Y, Pardo A, Koschel D, Bonella F, Spagnolo P, et al. Hypersensitivity pneumonitis. Nat Rev Dis Primers . 2020;6:65. - PubMed
-
- Selman M, Pardo A, King TE., Jr Hypersensitivity pneumonitis: insights in diagnosis and pathobiology. Am J Respir Crit Care Med . 2012;186:314–324. - PubMed
MeSH terms
Grants and funding
- R01 HL141852/HL/NHLBI NIH HHS/United States
- R21 LM012884/LM/NLM NIH HHS/United States
- R01 HL152677/HL/NHLBI NIH HHS/United States
- K23 HL146942/HL/NHLBI NIH HHS/United States
- R01 HL127349/HL/NHLBI NIH HHS/United States
- R01 LM014087/LM/NLM NIH HHS/United States
- T32 GM086287/GM/NIGMS NIH HHS/United States
- F30 HL162459/HL/NHLBI NIH HHS/United States
- Pulmonary Fibrosis Foundation/United States
- 1K08HL151970-01/HL/NHLBI NIH HHS/United States
- K08 HL151970/HL/NHLBI NIH HHS/United States
- Three Lakes Foundation/United States
- R21 HL161723/HL/NHLBI NIH HHS/United States
- R01 HL163984/HL/NHLBI NIH HHS/United States
- T32GM086287/GM/NIGMS NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- Veracyte/United States
- U01 HL145567/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

