Exploratory multi-omics analysis reveals host-microbe interactions associated with disease severity in psoriatic skin
- PMID: 38924840
- PMCID: PMC11259698
- DOI: 10.1016/j.ebiom.2024.105222
Exploratory multi-omics analysis reveals host-microbe interactions associated with disease severity in psoriatic skin
Abstract
Background: Psoriasis (Pso) is a chronic inflammatory skin disease that poses both physical and psychological challenges. Dysbiosis of the skin microbiome has been implicated in Pso, yet a comprehensive multi-omics analysis of host-microbe interactions is still lacking. To bridge this gap, we conducted an exploratory study by adopting the integrated approach that combines whole metagenomic shotgun sequencing with skin transcriptomics.
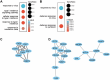
Methods: This was a cross-sectional study, adult patients with plaque-type Psoriasis (Pso) and healthy volunteers were included. Skin microbiota samples and biopsies were collected from both lesional and non-lesional skin areas on the lower back. Weighted Gene Correlation Network Analysis (WGCNA) was employed for co-expression network analysis, and cell deconvolution was conducted to estimate cell fractions. Taxonomic and functional features of the microbiome were identified using whole metagenomic shotgun sequencing. Association between host genes and microbes was analyzed using Spearman correlation.
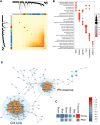
Findings: Host anti-viral responses and interferon-related networks were identified and correlated with the severity of psoriasis. The skin microbiome showed a greater prevalence of Corynebacterium simulans in the PASI severe-moderate groups, which correlated with interferon-induced host genes. Two distinct psoriatic clusters with varying disease severities were identified. Variations in the expression of cell apoptosis-associated antimicrobial peptides (AMPs) and microbial aerobic respiration I pathway may partly account for these differences in disease severity.
Interpretation: Our multi-omics analysis revealed for the first time anti-viral responses and the presence of C. simulans associated with psoriasis severity. It also identified two psoriatic subtypes with distinct AMP and metabolic pathway expression. Our study provides new insights into understanding the host-microbe interaction in psoriasis and lays the groundwork for developing subtype-specific strategies for managing this chronic skin disease.
Funding: The research has received funding from the FP7 (MAARS-Grant 261366) and the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 821511 (BIOMAP). The JU receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA. This publication reflects only the author's view and the JU is not responsible for any use that may be made of the information it contains. GAM was supported by a scholarship provided by CAPES-PRINT, financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES (Brazilian Government Agency). The authors thank all patients who participated in our study.
Keywords: Microbiome; Multi-omics; Psoriasis; Skin; Transcriptome.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of interests Prof. Homey declared a conflict of interests as described in the ICMJE DISCLOSURE FORM. All the other authors declare no competing interests.
Figures





References
-
- Parisi R., Symmons D.P., Griffiths C.E., et al. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol. 2013;133(2):377–385. - PubMed
-
- Elder J.T., Bruce A.T., Gudjonsson J.E., et al. Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol. 2010;130(5):1213–1226. - PubMed
-
- Pompili M., Bonanni L., Gualtieri F., Trovini G., Persechino S., Baldessarini R.J. Suicidal risks with psoriasis and atopic dermatitis: systematic review and meta-analysis. J Psychosom Res. 2021;141 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

