Characterization of Mice Carrying a Neurodevelopmental Disease-Associated GluN2B(L825V) Variant
- PMID: 38926089
- PMCID: PMC11293445
- DOI: 10.1523/JNEUROSCI.2291-23.2024
Characterization of Mice Carrying a Neurodevelopmental Disease-Associated GluN2B(L825V) Variant
Abstract
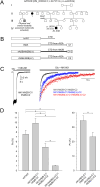
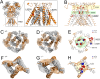
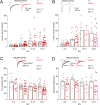
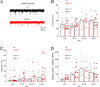
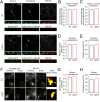
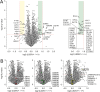
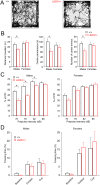
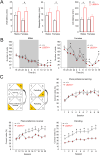
N-Methyl-d-aspartate receptors (NMDARs), encoded by GRIN genes, are ionotropic glutamate receptors playing a critical role in synaptic transmission, plasticity, and synapse development. Genome sequence analyses have identified variants in GRIN genes in patients with neurodevelopmental disorders, but the underlying disease mechanisms are not well understood. Here, we have created and evaluated a transgenic mouse line carrying a missense variant Grin2bL825V , corresponding to a de novo GRIN2B variant encoding GluN2B(L825V) found in a patient with intellectual disability (ID) and autism spectrum disorder (ASD). We used HEK293T cells expressing recombinant receptors and primary hippocampal neurons prepared from heterozygous Grin2bL825V/+ (L825V/+) and wild-type (WT) Grin2b+/+ (+/+) male and female mice to assess the functional impact of the variant. Whole-cell NMDAR currents were reduced in neurons from L825V/+ compared with +/+ mice. The peak amplitude of NMDAR-mediated evoked excitatory postsynaptic currents (NMDAR-eEPSCs) was unchanged, but NMDAR-eEPSCs in L825V/+ neurons had faster deactivation compared with +/+ neurons and were less sensitive to a GluN2B-selective antagonist ifenprodil. Together, these results suggest a decreased functional contribution of GluN2B subunits to synaptic NMDAR currents in hippocampal neurons from L825V/+ mice. The analysis of the GluN2B(L825V) subunit surface expression and synaptic localization revealed no differences compared with WT GluN2B. Behavioral testing of mice of both sexes demonstrated hypoactivity, anxiety, and impaired sensorimotor gating in the L825V/+ strain, particularly affecting males, as well as cognitive symptoms. The heterozygous L825V/+ mouse offers a clinically relevant model of GRIN2B-related ID/ASD, and our results suggest synaptic-level functional changes that may contribute to neurodevelopmental pathology.
Keywords: GluN2B; NMDA receptors; autism spectrum disorder; mouse model; synaptic transmission.
Copyright © 2024 the authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Akashi K, Kakizaki T, Kamiya H, Fukaya M, Yamasaki M, Abe M, Natsume R, Watanabe M, Sakimura K (2009) NMDA receptor GluN2B (GluR epsilon 2/NR2B) subunit is crucial for channel function, postsynaptic macromolecular organization, and actin cytoskeleton at hippocampal CA3 synapses. J Neurosci 29:10869–10882. 10.1523/JNEUROSCI.5531-08.2009 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
