Evolution of STAT2 resistance to flavivirus NS5 occurred multiple times despite genetic constraints
- PMID: 38926343
- PMCID: PMC11208600
- DOI: 10.1038/s41467-024-49758-0
Evolution of STAT2 resistance to flavivirus NS5 occurred multiple times despite genetic constraints
Abstract
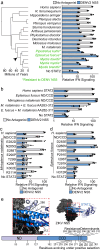
Zika and dengue virus nonstructural protein 5 antagonism of STAT2, a critical interferon signaling transcription factor, to suppress the host interferon response is required for viremia and pathogenesis in a vertebrate host. This affects viral species tropism, as mouse STAT2 resistance renders only immunocompromised or humanized STAT2 mice infectable. Here, we explore how STAT2 evolution impacts antagonism. By measuring the susceptibility of 38 diverse STAT2 proteins, we demonstrate that resistance arose numerous times in mammalian evolution. In four species, resistance requires distinct sets of multiple amino acid changes that often individually disrupt STAT2 signaling. This reflects an evolutionary ridge where progressive resistance is balanced by the need to maintain STAT2 function. Furthermore, resistance may come with a fitness cost, as resistance that arose early in lemur evolution was subsequently lost in some lemur lineages. These findings underscore that while it is possible to evolve resistance to antagonism, complex evolutionary trajectories are required to avoid detrimental host fitness consequences.
© 2024. The Author(s).
Conflict of interest statement
Adolfo García-Sastre has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, Pfizer and Prosetta, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and Astrazeneca. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- T32 AI007647/AI/NIAID NIH HHS/United States
- GM134936/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 AI083203/AI/NIAID NIH HHS/United States
- R35 GM134936/GM/NIGMS NIH HHS/United States
- AI007647/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

