Microglia contribute to neuronal synchrony despite endogenous ATP-related phenotypic transformation in acute mouse brain slices
- PMID: 38926390
- PMCID: PMC11208608
- DOI: 10.1038/s41467-024-49773-1
Microglia contribute to neuronal synchrony despite endogenous ATP-related phenotypic transformation in acute mouse brain slices
Abstract
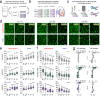
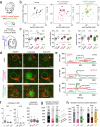
Acute brain slices represent a workhorse model for studying the central nervous system (CNS) from nanoscale events to complex circuits. While slice preparation inherently involves tissue damage, it is unclear how microglia, the main immune cells and damage sensors of the CNS react to this injury and shape neuronal activity ex vivo. To this end, we investigated microglial phenotypes and contribution to network organization and functioning in acute brain slices. We reveal time-dependent microglial phenotype changes influenced by complex extracellular ATP dynamics through P2Y12R and CX3CR1 signalling, which is sustained for hours in ex vivo mouse brain slices. Downregulation of P2Y12R and changes of microglia-neuron interactions occur in line with alterations in the number of excitatory and inhibitory synapses over time. Importantly, functional microglia modulate synapse sprouting, while microglial dysfunction results in markedly impaired ripple activity both ex vivo and in vivo. Collectively, our data suggest that microglia are modulators of complex neuronal networks with important roles to maintain neuronal network integrity and activity. We suggest that slice preparation can be used to model time-dependent changes of microglia-neuron interactions to reveal how microglia shape neuronal circuits in physiological and pathological conditions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Andersen P. Brain slices—a neurobiological tool of increasing usefulness. Trends Neurosci. 1981;4:53–56. doi: 10.1016/0166-2236(81)90019-9. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

