An intermediate Rb-E2F activity state safeguards proliferation commitment
- PMID: 38926571
- PMCID: PMC11236703
- DOI: 10.1038/s41586-024-07554-2
An intermediate Rb-E2F activity state safeguards proliferation commitment
Abstract
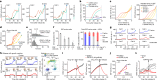
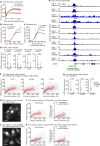
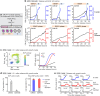
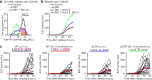
Tissue repair, immune defence and cancer progression rely on a vital cellular decision between quiescence and proliferation1,2. Mammalian cells proliferate by triggering a positive feedback mechanism3,4. The transcription factor E2F activates cyclin-dependent kinase 2 (CDK2), which in turn phosphorylates and inactivates the E2F inhibitor protein retinoblastoma (Rb). This action further increases E2F activity to express genes needed for proliferation. Given that positive feedback can inadvertently amplify small signals, understanding how cells keep this positive feedback in check remains a puzzle. Here we measured E2F and CDK2 signal changes in single cells and found that the positive feedback mechanism engages only late in G1 phase. Cells spend variable and often extended times in a reversible state of intermediate E2F activity before committing to proliferate. This intermediate E2F activity is proportional to the amount of phosphorylation of a conserved T373 residue in Rb that is mediated by CDK2 or CDK4/CDK6. Such T373-phosphorylated Rb remains bound on chromatin but dissociates from it once Rb is hyperphosphorylated at many sites, which fully activates E2F. The preferential initial phosphorylation of T373 can be explained by its relatively slower rate of dephosphorylation. Together, our study identifies a primed state of intermediate E2F activation whereby cells sense external and internal signals and decide whether to reverse and exit to quiescence or trigger the positive feedback mechanism that initiates cell proliferation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
-
- Xia, H. et al. Tissue repair and regeneration with endogenous stem cells. Nat. Rev. Mater.3, 174–193 (2018).10.1038/s41578-018-0027-6 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

