Structural mechanism of bridge RNA-guided recombination
- PMID: 38926616
- PMCID: PMC11208158
- DOI: 10.1038/s41586-024-07570-2
Structural mechanism of bridge RNA-guided recombination
Abstract
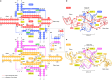
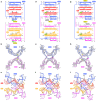
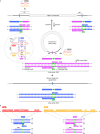
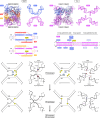
Insertion sequence (IS) elements are the simplest autonomous transposable elements found in prokaryotic genomes1. We recently discovered that IS110 family elements encode a recombinase and a non-coding bridge RNA (bRNA) that confers modular specificity for target DNA and donor DNA through two programmable loops2. Here we report the cryo-electron microscopy structures of the IS110 recombinase in complex with its bRNA, target DNA and donor DNA in three different stages of the recombination reaction cycle. The IS110 synaptic complex comprises two recombinase dimers, one of which houses the target-binding loop of the bRNA and binds to target DNA, whereas the other coordinates the bRNA donor-binding loop and donor DNA. We uncovered the formation of a composite RuvC-Tnp active site that spans the two dimers, positioning the catalytic serine residues adjacent to the recombination sites in both target and donor DNA. A comparison of the three structures revealed that (1) the top strands of target and donor DNA are cleaved at the composite active sites to form covalent 5'-phosphoserine intermediates, (2) the cleaved DNA strands are exchanged and religated to create a Holliday junction intermediate, and (3) this intermediate is subsequently resolved by cleavage of the bottom strands. Overall, this study reveals the mechanism by which a bispecific RNA confers target and donor DNA specificity to IS110 recombinases for programmable DNA recombination.
© 2024. The Author(s).
Conflict of interest statement
P.D.H. acknowledges outside interests in Stylus Medicine, Spotlight Therapeutics, Circle Labs, Arbor Biosciences, Varda Space, Vial Health, Serotiny and LabSimply, where he holds various roles as co-founder, director, scientific advisory board member or consultant. M.G.D. acknowledges outside interest in Stylus Medicine. M.H., N.T.P., M.G.D., J.S.A., S.K., P.D.H. and H.N. are listed as co-inventors on patent applications related to the structure-based engineering of IS110 systems, filed by Arc Institute, The University of Tokyo and the University of California. All other authors declare no competing interests.
Figures











References
-
- Siguier, P., Gourbeyre, E., Varani, A., Ton-Hoang, B. & Chandler, M. Everyman’s guide to bacterial insertion sequences. Microbiol. Spectr.3, MDNA3-0030-2014 (2015). - PubMed
-
- Curcio, M. J. & Derbyshire, K. M. The outs and ins of transposition: from Mu to Kangaroo. Nat. Rev. Mol. Cell Biol.4, 865–877 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

