FADS1/2 control lipid metabolism and ferroptosis susceptibility in triple-negative breast cancer
- PMID: 38926633
- PMCID: PMC11251055
- DOI: 10.1038/s44321-024-00090-6
FADS1/2 control lipid metabolism and ferroptosis susceptibility in triple-negative breast cancer
Abstract
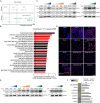
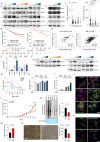
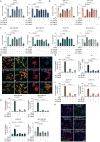
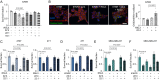
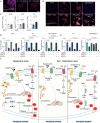
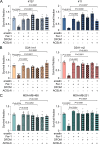
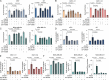
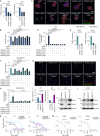
Triple-negative breast cancer (TNBC) has limited therapeutic options, is highly metastatic and characterized by early recurrence. Lipid metabolism is generally deregulated in TNBC and might reveal vulnerabilities to be targeted or used as biomarkers with clinical value. Ferroptosis is a type of cell death caused by iron-dependent lipid peroxidation which is facilitated by the presence of polyunsaturated fatty acids (PUFA). Here we identify fatty acid desaturases 1 and 2 (FADS1/2), which are responsible for PUFA biosynthesis, to be highly expressed in a subset of TNBC with a poorer prognosis. Lipidomic analysis, coupled with functional metabolic assays, showed that FADS1/2 high-expressing TNBC are susceptible to ferroptosis-inducing agents and that targeting FADS1/2 by both genetic interference and pharmacological approach renders those tumors ferroptosis-resistant while unbalancing PUFA/MUFA ratio by the supplementation of exogenous PUFA sensitizes resistant tumors to ferroptosis induction. Last, inhibiting lipid droplet (LD) formation and turnover suppresses the buffering capacity of LD and potentiates iron-dependent cell death. These findings have been validated in vitro and in vivo in mouse- and human-derived clinically relevant models and in a retrospective cohort of TNBC patients.
Keywords: Desaturases; Ferroptosis; Lipid Droplets; Lipid Metabolism; Polyunsaturated Fatty Acids.
© 2024. The Author(s).
Conflict of interest statement
IM declares consultant honoraria for Eli Lilly, Novartis, Seagen, Istituto Gentili, Roche, Pfizer, Ipsen, and Pierre Fabre. All the other authors declare no competing interests.
Figures




References
-
- Amaro A, Angelini G, Mirisola V, Esposito AI, Reverberi D, Matis S, Maffei M, Giaretti W, Viale M, Gangemi R et al (2016) A highly invasive subpopulation of MDA-MB-231 breast cancer cells shows accelerated growth, differential chemoresistance, features of apocrine tumors and reduced tumorigenicity in vivo. Oncotarget 7:68803–68820 - PMC - PubMed
-
- Bacci M, Lorito N, Smiriglia A, Morandi A (2021) Fat and furious: lipid metabolism in antitumoral therapy response and resistance. Trends Cancer 7:198–213 - PubMed
-
- Bacci M, Lorito N, Smiriglia A, Subbiani A, Bonechi F, Comito G, Morriset L, El Botty R, Benelli M, López-Velazco JI et al (2024) Acetyl-CoA carboxylase 1 controls a lipid droplet-peroxisome axis and is a vulnerability of endocrine-resistant ER. Sci Transl Med 16:eadf9874 - PubMed
-
- Brueffer C, Vallon-Christersson J, Grabau D, Ehinger A, Häkkinen J, Hegardt C, Malina J, Chen Y, Bendahl PO, Manjer J et al (2018) Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden cancerome analysis network-breast initiative. JCO Precis Oncol 2:PO.17.00135 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

