Integration of Proteomic and Metabolomic Data Reveals the Lipid Metabolism Disorder in the Liver of Rats Exposed to Simulated Microgravity
- PMID: 38927087
- PMCID: PMC11201887
- DOI: 10.3390/biom14060682
Integration of Proteomic and Metabolomic Data Reveals the Lipid Metabolism Disorder in the Liver of Rats Exposed to Simulated Microgravity
Abstract
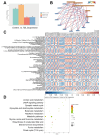
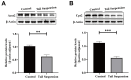
Long-term exposure to microgravity is considered to cause liver lipid accumulation, thereby increasing the risk of non-alcoholic fatty liver disease (NAFLD) among astronauts. However, the reasons for this persistence of symptoms remain insufficiently investigated. In this study, we used tandem mass tag (TMT)-based quantitative proteomics techniques, as well as non-targeted metabolomics techniques based on liquid chromatography-tandem mass spectrometry (LC-MS/MS), to comprehensively analyse the relative expression levels of proteins and the abundance of metabolites associated with lipid accumulation in rat liver tissues under simulated microgravity conditions. The differential analysis revealed 63 proteins and 150 metabolites between the simulated microgravity group and the control group. By integrating differentially expressed proteins and metabolites and performing pathway enrichment analysis, we revealed the dysregulation of major metabolic pathways under simulated microgravity conditions, including the biosynthesis of unsaturated fatty acids, linoleic acid metabolism, steroid hormone biosynthesis and butanoate metabolism, indicating disrupted liver metabolism in rats due to weightlessness. Finally, we examined differentially expressed proteins associated with lipid metabolism in the liver of rats exposed to stimulated microgravity. These findings contribute to identifying the key molecules affected by microgravity and could guide the design of rational nutritional or pharmacological countermeasures for astronauts.
Keywords: lipid metabolism; metabolomics; proteomics; simulated microgravity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Afshinnekoo E., Scott R.T., MacKay M.J., Pariset E., Cekanaviciute E., Barker R., Gilroy S., Hassane D., Smith S.M., Zwart S.R., et al. Fundamental Biological Features of Spaceflight: Advancing the Field to Enable Deep-Space Exploration. Cell. 2020;183:1162–1184. doi: 10.1016/j.cell.2020.10.050. - DOI - PMC - PubMed
-
- Radugina E.A., Almeida E.A.C., Blaber E., Poplinskaya V.A., Markitantova Y.V., Grigoryan E.N. Exposure to microgravity for 30 days onboard Bion M1 caused muscle atrophy and impaired regeneration in murine femoral Quadriceps. Life Sci. Space Res. 2018;16:18–25. doi: 10.1016/j.lssr.2017.08.005. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

