Accurate Early Detection and EGFR Mutation Status Prediction of Lung Cancer Using Plasma cfDNA Coverage Patterns: A Proof-of-Concept Study
- PMID: 38927119
- PMCID: PMC11202186
- DOI: 10.3390/biom14060716
Accurate Early Detection and EGFR Mutation Status Prediction of Lung Cancer Using Plasma cfDNA Coverage Patterns: A Proof-of-Concept Study
Abstract
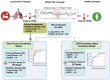
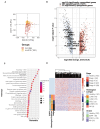
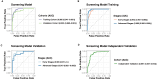
Lung cancer is a major global health concern with a low survival rate, often due to late-stage diagnosis. Liquid biopsy offers a non-invasive approach to cancer detection and monitoring, utilizing various features of circulating cell-free DNA (cfDNA). In this study, we established two models based on cfDNA coverage patterns at the transcription start sites (TSSs) from 6X whole-genome sequencing: an Early Cancer Screening Model and an EGFR mutation status prediction model. The Early Cancer Screening Model showed encouraging prediction ability, especially for early-stage lung cancer. The EGFR mutation status prediction model exhibited high accuracy in distinguishing between EGFR-positive and wild-type cases. Additionally, cfDNA coverage patterns at TSSs also reflect gene expression patterns at the pathway level in lung cancer patients. These findings demonstrate the potential applications of cfDNA coverage patterns at TSSs in early cancer screening and in cancer subtyping.
Keywords: EGFR mutation status prediction; cfDNA; coverage patterns at the transcription start sites; early cancer screening; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Raman L., Van der Linden M., Van der Eecken K., Vermaelen K., Demedts I., Surmont V., Himpe U., Dedeurwaerdere F., Ferdinande L., Lievens Y. Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma. Genome Med. 2020;12:35. doi: 10.1186/s13073-020-00735-4. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

