CBP/P300 Inhibition Impairs CD4+ T Cell Activation: Implications for Autoimmune Disorders
- PMID: 38927552
- PMCID: PMC11202127
- DOI: 10.3390/biomedicines12061344
CBP/P300 Inhibition Impairs CD4+ T Cell Activation: Implications for Autoimmune Disorders
Abstract
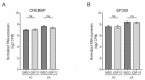
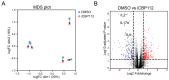
T cell activation is critical for an effective immune response against pathogens. However, dysregulation contributes to the pathogenesis of autoimmune diseases, including Juvenile Idiopathic Arthritis (JIA). The molecular mechanisms underlying T cell activation are still incompletely understood. T cell activation promotes the acetylation of histone 3 at Lysine 27 (H3K27ac) at enhancer and promoter regions of proinflammatory cytokines, thereby increasing the expression of these genes which is essential for T cell function. Co-activators E1A binding protein P300 (P300) and CREB binding protein (CBP), collectively known as P300/CBP, are essential to facilitate H3K27 acetylation. Presently, the role of P300/CBP in human CD4+ T cells activation remains incompletely understood. To assess the function of P300/CBP in T cell activation and autoimmune disease, we utilized iCBP112, a selective inhibitor of P300/CBP, in T cells obtained from healthy controls and JIA patients. Treatment with iCBP112 suppressed T cell activation and cytokine signaling pathways, leading to reduced expression of many proinflammatory cytokines, including IL-2, IFN-γ, IL-4, and IL-17A. Moreover, P300/CBP inhibition in T cells derived from the inflamed synovium of JIA patients resulted in decreased expression of similar pathways and preferentially suppressed the expression of disease-associated genes. This study underscores the regulatory role of P300/CBP in regulating gene expression during T cell activation while offering potential insights into the pathogenesis of autoimmune diseases. Our findings indicate that P300/CBP inhibition could potentially be leveraged for the treatment of autoimmune diseases such as JIA in the future.
Keywords: H3K27ac; JIA; P300/CBP; T cell activation; autoimmune diseases; cytokines; iCBP112.
Conflict of interest statement
David Tough and Rab Prinjha are employees and shareholders of GSK. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Figures






References
-
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

