Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model
- PMID: 38927934
- PMCID: PMC11201561
- DOI: 10.3390/cancers16122229
Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model
Abstract
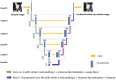
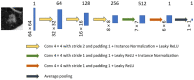
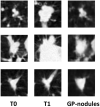
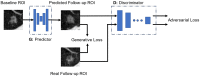
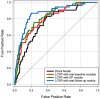
Early diagnosis of lung cancer can significantly improve patient outcomes. We developed a Growth Predictive model based on the Wasserstein Generative Adversarial Network framework (GP-WGAN) to predict the nodule growth patterns in the follow-up LDCT scans. The GP-WGAN was trained with a training set (N = 776) containing 1121 pairs of nodule images with about 1-year intervals and deployed to an independent test set of 450 nodules on baseline LDCT scans to predict nodule images (GP-nodules) in their 1-year follow-up scans. The 450 GP-nodules were finally classified as malignant or benign by a lung cancer risk prediction (LCRP) model, achieving a test AUC of 0.827 ± 0.028, which was comparable to the AUC of 0.862 ± 0.028 achieved by the same LCRP model classifying real follow-up nodule images (p = 0.071). The net reclassification index yielded consistent outcomes (NRI = 0.04; p = 0.62). Other baseline methods, including Lung-RADS and the Brock model, achieved significantly lower performance (p < 0.05). The results demonstrated that the GP-nodules predicted by our GP-WGAN model achieved comparable performance with the nodules in the real follow-up scans for lung cancer diagnosis, indicating the potential to detect lung cancer earlier when coupled with accelerated clinical management versus the current approach of waiting until the next screening exam.
Keywords: deep learning; early diagnosis; generative AI; lung cancer; nodule growth prediction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Cancer Stat Facts: Lung and Bronchus Cancer. [(accessed on 1 September 2022)]; Available online: https://seer.cancer.gov/statfacts/html/lungb.html.
-
- Howlader N., Noone A.M., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. SEER Cancer Statistics Review, 1975–2018. [(accessed on 1 September 2022)]; Available online: https://seer.cancer.gov/archive/csr/1975_2018/index.html.
-
- Tao G., Zhu L., Chen Q., Yin L., Li Y., Yang J., Ni B., Zhang Z., Koo C.W., Patil P.D. Prediction of future imagery of lung nodule as growth modeling with follow-up computed tomography scans using deep learning: A retrospective cohort study. Transl. Lung Cancer Res. 2022;11:250. doi: 10.21037/tlcr-22-59. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

