Detection and Characterisation of SARS-CoV-2 in Eastern Province of Zambia: A Retrospective Genomic Surveillance Study
- PMID: 38928045
- PMCID: PMC11203853
- DOI: 10.3390/ijms25126338
Detection and Characterisation of SARS-CoV-2 in Eastern Province of Zambia: A Retrospective Genomic Surveillance Study
Abstract
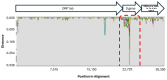
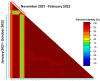
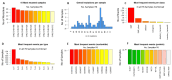
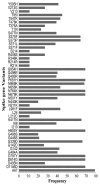
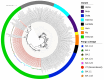
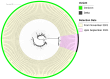
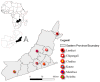
Mutations have driven the evolution and development of new variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with potential implications for increased transmissibility, disease severity and vaccine escape among others. Genome sequencing is a technique that allows scientists to read the genetic code of an organism and has become a powerful tool for studying emerging infectious diseases. Here, we conducted a cross-sectional study in selected districts of the Eastern Province of Zambia, from November 2021 to February 2022. We analyzed SARS-CoV-2 samples (n = 76) using high-throughput sequencing. A total of 4097 mutations were identified in 69 SARS-CoV-2 genomes with 47% (1925/4097) of the mutations occurring in the spike protein. We identified 83 unique amino acid mutations in the spike protein of the seven Omicron sublineages (BA.1, BA.1.1, BA.1.14, BA.1.18, BA.1.21, BA.2, BA.2.23 and XT). Of these, 43.4% (36/83) were present in the receptor binding domain, while 14.5% (12/83) were in the receptor binding motif. While we identified a potential recombinant XT strain, the highly transmissible BA.2 sublineage was more predominant (40.8%). We observed the substitution of other variants with the Omicron strain in the Eastern Province. This work shows the importance of pandemic preparedness and the need to monitor disease in the general population.
Keywords: Eastern Province; Omicron variant; SAR-CoV-2 mutations; SARS-CoV-2; Zambia; genomic surveillance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
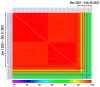
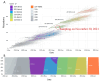
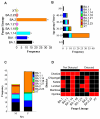
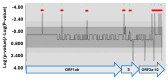
Similar articles
-
Genomic Surveillance of SARS-CoV-2 in the Southern Province of Zambia: Detection and Characterization of Alpha, Beta, Delta, and Omicron Variants of Concern.Viruses. 2022 Aug 24;14(9):1865. doi: 10.3390/v14091865. Viruses. 2022. PMID: 36146671 Free PMC article.
-
Genetic diversity and evolutionary dynamics of the Omicron variant of SARS-CoV-2 in Morocco.Pathog Glob Health. 2024 May;118(3):241-252. doi: 10.1080/20477724.2023.2250942. Epub 2023 Aug 27. Pathog Glob Health. 2024. PMID: 37635364 Free PMC article.
-
Dominance of SARS-CoV-2 Delta AY.33 sublineage and Omicron BA.1.1 sublineage in Erbil City/Kurdistan Region of Iraq.Cell Mol Biol (Noisy-le-grand). 2024 Nov 24;70(10):83-90. doi: 10.14715/cmb/2024.70.10.12. Cell Mol Biol (Noisy-le-grand). 2024. PMID: 39605120
-
The outbreak of SARS-CoV-2 Omicron lineages, immune escape, and vaccine effectivity.J Med Virol. 2023 Jan;95(1):e28138. doi: 10.1002/jmv.28138. Epub 2022 Sep 21. J Med Virol. 2023. PMID: 36097349 Free PMC article. Review.
-
JN.1 variant in enduring COVID-19 pandemic: is it a variety of interest (VoI) or variety of concern (VoC)?Horm Mol Biol Clin Investig. 2024 Apr 17;45(2):49-53. doi: 10.1515/hmbci-2023-0088. eCollection 2024 Jun 1. Horm Mol Biol Clin Investig. 2024. PMID: 38622986 Review.
Cited by
-
Wastewater Surveillance of SARS-CoV-2 in Zambia: An Early Warning Tool.Int J Mol Sci. 2024 Aug 14;25(16):8839. doi: 10.3390/ijms25168839. Int J Mol Sci. 2024. PMID: 39201525 Free PMC article.
References
-
- Santaniello A., Perruolo G., Cristiano S., Agognon A.L., Cabaro S., Amato A., Dipineto L., Borrelli L., Formisano P., Fioretti A., et al. SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review. Microorganisms. 2023;11:514. doi: 10.3390/microorganisms11020514. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

