Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma
- PMID: 38928223
- PMCID: PMC11204065
- DOI: 10.3390/ijms25126518
Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma
Abstract
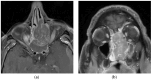
Mutations affecting codon 172 of the isocitrate dehydrogenase 2 (IDH2) gene define a subgroup of sinonasal undifferentiated carcinomas (SNUCs) with a relatively favorable prognosis and a globally hypermethylated phenotype. They are also recurrent (along with IDH1 mutations) in gliomas, acute myeloid leukemia, and intrahepatic cholangiocarcinoma. Commonly reported mutations, all associated with aberrant IDH2 enzymatic activity, include R172K, R172S, R172T, R172G, and R172M. We present a case of SNUC with a never-before-described IDH2 mutation, R172A. Our report compares the methylation pattern of our sample to other cases from the Gene Expression Omnibus database. Hierarchical clustering suggests a strong association between our sample and other IDH-mutant SNUCs and a clear distinction between sinonasal normal tissues and tumors. Principal component analysis (PCA), using 100 principal components explaining 94.5% of the variance, showed the position of our sample to be within 1.02 standard deviation of the other IDH-mutant SNUCs. A molecular modeling analysis of the IDH2 R172A versus other R172 variants provides a structural explanation to how they affect the protein active site. Our findings thus suggest that the R172A mutation in IDH2 confers a gain of function similar to other R172 mutations in IDH2, resulting in a similar hypermethylated profile.
Keywords: IDH2 mutation; methylation analysis; molecular modeling.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




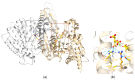




References
-
- Showalter M.R., Hatakeyama J., Cajka T., VanderVorst K., Carraway K.L., Fiehn O., Reproducibility Project: Cancer Biology Replication Study: The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. eLife. 2017;6:e26030. doi: 10.7554/eLife.26030. - DOI - PMC - PubMed
-
- Nakazawa S., Sakata K.I., Liang S., Yoshikawa K., Iizasa H., Tada M., Hamada J.I., Kashiwazaki H., Kitagawa Y., Yamazaki Y. Dominant-negative p53 mutant R248Q increases the motile and invasive activities of oral squamous cell carcinoma cells. Biomed Res. 2019;40:37–49. doi: 10.2220/biomedres.40.37. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

