Exploring the Potential Role of Oligodendrocyte-Associated PIP4K2A in Alzheimer's Disease Complicated with Type 2 Diabetes Mellitus via Multi-Omic Analysis
- PMID: 38928345
- PMCID: PMC11204139
- DOI: 10.3390/ijms25126640
Exploring the Potential Role of Oligodendrocyte-Associated PIP4K2A in Alzheimer's Disease Complicated with Type 2 Diabetes Mellitus via Multi-Omic Analysis
Abstract
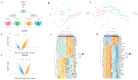
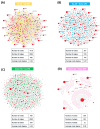
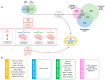
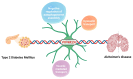
Alzheimer's disease (AD) and type 2 diabetes mellitus (T2DM) are two common diseases that affect the elderly population worldwide. The identification of common genes associated with AD and T2DM holds promise for potential biomarkers and intriguing pathogenesis of these two complicated diseases. This study utilized a comprehensive approach by integrating transcriptome data from multiple cohorts, encompassing both AD and T2DM. The analysis incorporated various data types, including blood and tissue samples as well as single-cell datasets, allowing for a detailed assessment of gene expression patterns. From the brain region-specific single-cell analysis, PIP4K2A, which encodes phosphatidylinositol-5-phosphate 4-kinase type 2 alpha, was found to be expressed mainly in oligodendrocytes compared to other cell types. Elevated levels of PIP4K2A in AD and T2DM patients' blood were found to be associated with key cellular processes such as vesicle-mediated transport, negative regulation of autophagosome assembly, and cytosolic transport. The identification of PIP4K2A's potential roles in the cellular processes of AD and T2DM offers valuable insights into the development of biomarkers for diagnosis and therapy, especially in the complication of these two diseases.
Keywords: Alzheimer’s disease; PIP4K2A; bioinformatics; biomarker; single-cell sequencing; type 2 diabetes mellitus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Integration of Single-Cell and Spatial Transcriptomic Data Reveals Spatial Architecture and Potential Biomarkers in Alzheimer's Disease.Mol Neurobiol. 2025 May;62(5):5395-5412. doi: 10.1007/s12035-024-04617-3. Epub 2024 Nov 15. Mol Neurobiol. 2025. PMID: 39543008
-
Exploring Shared Pathogenesis of Alzheimer's Disease and Type 2 Diabetes Mellitus via Co-expression Networks Analysis.Curr Alzheimer Res. 2020;17(6):566-575. doi: 10.2174/1567205017666200810164932. Curr Alzheimer Res. 2020. PMID: 32781959
-
Identification and Experimental Validation of Marker Genes between Diabetes and Alzheimer's Disease.Oxid Med Cell Longev. 2022 Aug 12;2022:8122532. doi: 10.1155/2022/8122532. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35996379 Free PMC article.
-
Alzheimer's disease and type 2 diabetes mellitus: A systematic review of proteomic studies.J Neurochem. 2021 Mar;156(6):753-776. doi: 10.1111/jnc.15166. Epub 2020 Oct 7. J Neurochem. 2021. PMID: 32909269
-
A Tale of Two Diseases: Exploring Mechanisms Linking Diabetes Mellitus with Alzheimer's Disease.J Alzheimers Dis. 2022;85(2):485-501. doi: 10.3233/JAD-210612. J Alzheimers Dis. 2022. PMID: 34842187 Review.
Cited by
-
Decoding neuroinflammation in Alzheimer's disease: a multi-omics and AI-driven perspective for precision medicine.Front Immunol. 2025 Jun 18;16:1616899. doi: 10.3389/fimmu.2025.1616899. eCollection 2025. Front Immunol. 2025. PMID: 40607399 Free PMC article. Review.
References
-
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

