Novel Ultrasound-Responsive Amyloid Formulation
- PMID: 38931443
- PMCID: PMC11206591
- DOI: 10.3390/ph17060777
Novel Ultrasound-Responsive Amyloid Formulation
Abstract
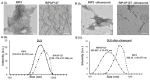
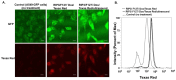
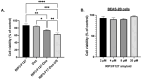
Amyloid aggregates have attracted significant interest in regard to diverse biomedical applications, particularly in the field of drug delivery. Here, we report novel amyloid aggregates based on a 12-amino-acid peptide from the amyloidogenic region of the receptor-interacting kinase 3 (RIP3) protein and a thermoresponsive triblock copolymer, namely, Pluronic F127 (RIP3/F127). Physicochemical characterization was performed to determine the aggregation size, morphology, and stimuli-responsive properties. The potential of the aggregates as a drug depot was assessed in lung cancer cells, using Doxorubicin (Dox) as a model drug. The results show that RIP3 and RIP3/F127 exhibit amyloidogenic properties. Further, the RIP3/F127 amyloids exhibited significant ultrasound-responsive properties compared to amyloid aggregates without Pluronic F127. Moreover, the RIP3/F127/Dox amyloid formulations that were subjected to ultrasound treatment exhibited greater toxicity to lung cancer cells compared to that of Dox alone at equal concentrations. Overall, the results from this proof-of-concept study show that amyloidogenic peptide aggregates with stimuli-responsive properties can be utilized as efficient drug delivery depots.
Keywords: RIP3; amyloid aggregates; drug delivery; thermoresponsive polymer.
Conflict of interest statement
The authors declare no conflicts of interest regarding the contents of the article.
Figures


References
-
- Li J., McQuade T., Siemer A.B., Napetschnig J., Moriwaki K., Hsiao Y.S., Damko E., Moquin D., Walz T., McDermott A., et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

