3-Chymotrypsin-like Protease (3CLpro) of SARS-CoV-2: Validation as a Molecular Target, Proposal of a Novel Catalytic Mechanism, and Inhibitors in Preclinical and Clinical Trials
- PMID: 38932137
- PMCID: PMC11209289
- DOI: 10.3390/v16060844
3-Chymotrypsin-like Protease (3CLpro) of SARS-CoV-2: Validation as a Molecular Target, Proposal of a Novel Catalytic Mechanism, and Inhibitors in Preclinical and Clinical Trials
Abstract
Proteases represent common targets in combating infectious diseases, including COVID-19. The 3-chymotrypsin-like protease (3CLpro) is a validated molecular target for COVID-19, and it is key for developing potent and selective inhibitors for inhibiting viral replication of SARS-CoV-2. In this review, we discuss structural relationships and diverse subsites of 3CLpro, shedding light on the pivotal role of dimerization and active site architecture in substrate recognition and catalysis. Our analysis of bioinformatics and other published studies motivated us to investigate a novel catalytic mechanism for the SARS-CoV-2 polyprotein cleavage by 3CLpro, centering on the triad mechanism involving His41-Cys145-Asp187 and its indispensable role in viral replication. Our hypothesis is that Asp187 may participate in modulating the pKa of the His41, in which catalytic histidine may act as an acid and/or a base in the catalytic mechanism. Recognizing Asp187 as a crucial component in the catalytic process underscores its significance as a fundamental pharmacophoric element in drug design. Next, we provide an overview of both covalent and non-covalent inhibitors, elucidating advancements in drug development observed in preclinical and clinical trials. By highlighting various chemical classes and their pharmacokinetic profiles, our review aims to guide future research directions toward the development of highly selective inhibitors, underscore the significance of 3CLpro as a validated therapeutic target, and propel the progression of drug candidates through preclinical and clinical phases.
Keywords: 3CLpro; SARS-CoV-2; novel mechanism of catalysis; preclinical and clinical trials; triad.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


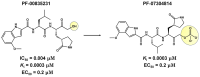

References
-
- Kar S., Ganguly M., Sen S. Lifting Scheme-Based Wavelet Transform Method for Improved Genomic Classification and Sequence Analysis of Coronavirus. Innov. Emerg. Technol. 2023;10:2350002. doi: 10.1142/S2737599423500020. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- 019/00195-2, 2020/04680-0, 2016/09047-8, 2022/08730-7/Fundação de Amparo à Pesquisa do Estado de São Paulo
- 88887.374931/2019-00/Coordenação de Aperfeicoamento de Pessoal de Nível Superior
- Rede Virus MCTI 0459/20/Instituto Nacional de Estudos e Pesquisa
- 2021/00070-5/Fundação de Amparo à Pesquisa do Estado de São Paulo
LinkOut - more resources
Full Text Sources
Miscellaneous

