Exploring Cannabinoids as Potential Inhibitors of SARS-CoV-2 Papain-like Protease: Insights from Computational Analysis and Molecular Dynamics Simulations
- PMID: 38932170
- PMCID: PMC11209085
- DOI: 10.3390/v16060878
Exploring Cannabinoids as Potential Inhibitors of SARS-CoV-2 Papain-like Protease: Insights from Computational Analysis and Molecular Dynamics Simulations
Abstract
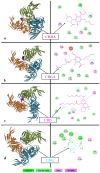
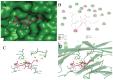
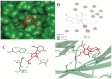
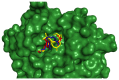
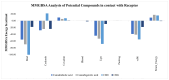
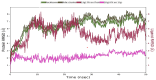
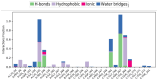
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has triggered a global COVID-19 pandemic, challenging healthcare systems worldwide. Effective therapeutic strategies against this novel coronavirus remain limited, underscoring the urgent need for innovative approaches. The present research investigates the potential of cannabis compounds as therapeutic agents against SARS-CoV-2 through their interaction with the virus's papain-like protease (PLpro) protein, a crucial element in viral replication and immune evasion. Computational methods, including molecular docking and molecular dynamics (MD) simulations, were employed to screen cannabis compounds against PLpro and analyze their binding mechanisms and interaction patterns. The results showed cannabinoids with binding affinities ranging from -6.1 kcal/mol to -4.6 kcal/mol, forming interactions with PLpro. Notably, Cannabigerolic and Cannabidiolic acids exhibited strong binding contacts with critical residues in PLpro's active region, indicating their potential as viral replication inhibitors. MD simulations revealed the dynamic behavior of cannabinoid-PLpro complexes, highlighting stable binding conformations and conformational changes over time. These findings shed light on the mechanisms underlying cannabis interaction with SARS-CoV-2 PLpro, aiding in the rational design of antiviral therapies. Future research will focus on experimental validation, optimizing binding affinity and selectivity, and preclinical assessments to develop effective treatments against COVID-19.
Keywords: PLpro; SARS-CoV-2; antiviral therapy; cannabis compounds; computational methods; molecular docking; molecular dynamics simulations; viral replication.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Disrupting protease and deubiquitinase activities of SARS-CoV-2 papain-like protease by natural and synthetic products discovered through multiple computational and biochemical approaches.Int J Biol Macromol. 2024 Oct;277(Pt 4):134476. doi: 10.1016/j.ijbiomac.2024.134476. Epub 2024 Aug 5. Int J Biol Macromol. 2024. PMID: 39111477
-
Discovery of SARS-CoV-2 papain-like protease inhibitors through machine learning and molecular simulation approaches.Drug Discov Ther. 2025 Jul 4;19(3):189-199. doi: 10.5582/ddt.2025.01034. Epub 2025 Jun 27. Drug Discov Ther. 2025. PMID: 40571587
-
Integrating Molecular Dynamics, Molecular Docking, and Machine Learning for Predicting SARS-CoV-2 Papain-like Protease Binders.Molecules. 2025 Jul 16;30(14):2985. doi: 10.3390/molecules30142985. Molecules. 2025. PMID: 40733251 Free PMC article.
-
Analysis of Structures of SARS-CoV-2 Papain-like Protease Bound with Ligands Unveils Structural Features for Inhibiting the Enzyme.Molecules. 2025 Jan 23;30(3):491. doi: 10.3390/molecules30030491. Molecules. 2025. PMID: 39942596 Free PMC article. Review.
-
Chalcone-amide, a privileged backbone for the design and development of selective SARS-CoV/SARS-CoV-2 papain-like protease inhibitors.Eur J Med Chem. 2022 Oct 5;240:114572. doi: 10.1016/j.ejmech.2022.114572. Epub 2022 Jul 3. Eur J Med Chem. 2022. PMID: 35797899 Free PMC article. Review.
Cited by
-
Anti-Inflammatory Effects of Cannabigerol In Vitro and In Vivo Are Mediated Through the JAK/STAT/NFκB Signaling Pathway.Cells. 2025 Jan 9;14(2):83. doi: 10.3390/cells14020083. Cells. 2025. PMID: 39851511 Free PMC article.
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

