Applying Flow Virometry to Study the HIV Envelope Glycoprotein and Differences Across HIV Model Systems
- PMID: 38932227
- PMCID: PMC11209363
- DOI: 10.3390/v16060935
Applying Flow Virometry to Study the HIV Envelope Glycoprotein and Differences Across HIV Model Systems
Abstract
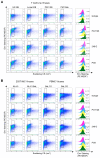
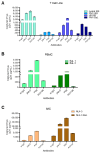
The HIV envelope glycoprotein (Env) is a trimeric protein that facilitates viral binding and fusion with target cells. As the sole viral protein on the HIV surface, Env is important both for immune responses to HIV and in vaccine designs. Targeting Env in clinical applications is challenging due to its heavy glycosylation, high genetic variability, conformational camouflage, and its low abundance on virions. Thus, there is a critical need to better understand this protein. Flow virometry (FV) is a useful methodology for phenotyping the virion surface in a high-throughput, single virion manner. To demonstrate the utility of FV to characterize Env, we stained HIV virions with a panel of 85 monoclonal antibodies targeting different regions of Env. A broad range of antibodies yielded robust staining of Env, with V3 antibodies showing the highest quantitative staining. A subset of antibodies tested in parallel on viruses produced in CD4+ T cell lines, HEK293T cells, and primary cells showed that the cellular model of virus production can impact Env detection. Finally, in addition to being able to highlight Env heterogeneity on virions, we show FV can sensitively detect differences in Env conformation when soluble CD4 is added to virions before staining.
Keywords: Env conformation; HIV Env; HIV trimer; calibrated flow virometry; gp120/gp41; human immunodeficiency virus (HIV); molecules of equivalent soluble fluorophore (MESF); nanoscale flow cytometry; neutralization; virion capture.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




Similar articles
-
Flow Cytometry Analysis of HIV-1 Env Conformations at the Surface of Infected Cells and Virions: Role of Nef, CD4, and SERINC5.J Virol. 2020 Feb 28;94(6):e01783-19. doi: 10.1128/JVI.01783-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31852789 Free PMC article.
-
Dual Pathways of Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Trafficking Modulate the Selective Exclusion of Uncleaved Oligomers from Virions.J Virol. 2021 Jan 13;95(3):e01369-20. doi: 10.1128/JVI.01369-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33148792 Free PMC article.
-
Characterization of the Human Immunodeficiency Virus (HIV-1) Envelope Glycoprotein Conformational States on Infectious Virus Particles.J Virol. 2023 Mar 30;97(3):e0185722. doi: 10.1128/jvi.01857-22. Epub 2023 Feb 23. J Virol. 2023. PMID: 36815832 Free PMC article.
-
HIV-1 Envelope Conformation, Allostery, and Dynamics.Viruses. 2021 May 7;13(5):852. doi: 10.3390/v13050852. Viruses. 2021. PMID: 34067073 Free PMC article. Review.
-
Fluorescence Microscopy of the HIV-1 Envelope.Viruses. 2020 Mar 21;12(3):348. doi: 10.3390/v12030348. Viruses. 2020. PMID: 32245254 Free PMC article. Review.
Cited by
-
Flow virometry: recent advancements, best practices, and future frontiers.J Virol. 2025 Feb 25;99(2):e0171724. doi: 10.1128/jvi.01717-24. Epub 2025 Jan 27. J Virol. 2025. PMID: 39868829 Free PMC article. Review.
-
Characterization of Nanobody Binding to Distinct Regions of the SARS-CoV-2 Spike Protein by Flow Virometry.Viruses. 2025 Apr 15;17(4):571. doi: 10.3390/v17040571. Viruses. 2025. PMID: 40285013 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

