The impact of high-IgE levels on metabolome and microbiome in experimental allergic enteritis
- PMID: 38932655
- PMCID: PMC11657046
- DOI: 10.1111/all.16202
The impact of high-IgE levels on metabolome and microbiome in experimental allergic enteritis
Abstract
Background: The pathological mechanism of the gastrointestinal forms of food allergies is less understood in comparison to other clinical phenotypes, such as asthma and anaphylaxis Importantly, high-IgE levels are a poor prognostic factor in gastrointestinal allergies.
Methods: This study investigated how high-IgE levels influence the development of intestinal inflammation and the metabolome in allergic enteritis (AE), using IgE knock-in (IgEki) mice expressing high levels of IgE. In addition, correlation of the altered metabolome with gut microbiome was analysed.
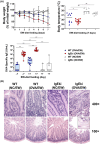
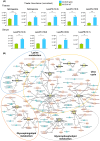
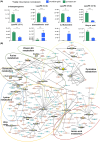
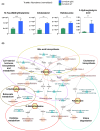
Results: Ovalbumin-sensitized and egg-white diet-fed (OVA/EW) BALB/c WT mice developed moderate AE, whereas OVA/EW IgEki mice induced more aggravated intestinal inflammation with enhanced eosinophil accumulation. Untargeted metabolomics detected the increased levels of N-tau-methylhistamine and 2,3-butanediol, and reduced levels of butyric acid in faeces and/or sera of OVA/EW IgEki mice, which was accompanied with reduced Clostridium and increased Lactobacillus at the genus level. Non-sensitized and egg-white diet-fed (NC/EW) WT mice did not exhibit any signs of AE, whereas NC/EW IgEki mice developed marginal degrees of AE. Compared to NC/EW WT mice, enhanced levels of lysophospholipids, sphinganine and sphingosine were detected in serum and faecal samples of NC/EW IgEki mice. In addition, several associations of altered metabolome with gut microbiome-for example Akkermansia with lysophosphatidylserine-were detected.
Conclusions: Our results suggest that high-IgE levels alter intestinal and systemic levels of endogenous and microbiota-associated metabolites in experimental AE. This study contributes to deepening the knowledge of molecular mechanisms for the development of AE and provides clues to advance diagnostic and therapeutic strategies of allergic diseases.
Keywords: IgE; food allergy; metabolomics; microbiome; microbiota; murine model.
© 2024 The Author(s). Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
Dr. Stefan Vieths reports personal honorarium from AAAAI as Associate Editor of J. Allergy Clin Immunol, non‐financial support from European Academy of Allergy and Clinical Immunology.
Figures



References
-
- Anvari S, Miller J, Yeh C‐Y, Davis CM. IgE‐mediated food allergy. Clin Rev Allergy Immunol. 2019;57:244‐260. - PubMed
-
- Ho MH‐K, Wong WH‐S, Chang C. Clinical spectrum of food allergies. A comprehensive review. Clin Rev Allergy Immunol. 2014;46:225‐240. - PubMed
-
- Burggraf M, Nakajima‐Adachi H, Hachimura S, et al. Oral tolerance induction does not resolve gastrointestinal inflammation in a mouse model of food allergy. Mol Nutr Food Res. 2011;55:1475‐1483. - PubMed
MeSH terms
Substances
Grants and funding
- Deutscher Akademischer Austausch Dienst (DAAD)
- (RD21/0002/0008)/FEDER 'Investing in your future' for the thematic network and co-operative research centres RICORS 'Red de Enfermedades Inflamatorias (REI)'
- Japan Society for the Promotion of Science (JSPS)
- PI19/00044/Instituto de Salud Carlos III
- PI17/01087/Instituto de Salud Carlos III
LinkOut - more resources
Full Text Sources

