The devil is in the detail: Environmental variables frequently used for habitat suitability modeling lack information for forest-dwelling bats in Germany
- PMID: 38932971
- PMCID: PMC11199919
- DOI: 10.1002/ece3.11571
The devil is in the detail: Environmental variables frequently used for habitat suitability modeling lack information for forest-dwelling bats in Germany
Abstract
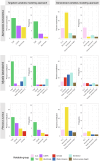
In response to the pressing challenges of the ongoing biodiversity crisis, the protection of endangered species and their habitats, as well as the monitoring of invasive species are crucial. Habitat suitability modeling (HSM) is often treated as the silver bullet to address these challenges, commonly relying on generic variables sourced from widely available datasets. However, for species with high habitat requirements, or for modeling the suitability of habitats within the geographic range of a species, variables at a coarse level of detail may fall short. Consequently, there is potential value in considering the incorporation of more targeted data, which may extend beyond readily available land cover and climate datasets. In this study, we investigate the impact of incorporating targeted land cover variables (specifically tree species composition) and vertical structure information (derived from LiDAR data) on HSM outcomes for three forest specialist bat species (Barbastella barbastellus, Myotis bechsteinii, and Plecotus auritus) in Rhineland-Palatinate, Germany, compared to commonly utilized environmental variables, such as generic land-cover classifications (e.g., Corine Land Cover) and climate variables (e.g., Bioclim). The integration of targeted variables enhanced the performance of habitat suitability models for all three bat species. Furthermore, our results showed a high difference in the distribution maps that resulted from using different levels of detail in environmental variables. This underscores the importance of making the effort to generate the appropriate variables, rather than simply relying on commonly used ones, and the necessity of exercising caution when using habitat models as a tool to inform conservation strategies and spatial planning efforts.
Keywords: bats; habitat suitability modeling; land cover/land use; nature conservation; spatialMaxent; species distribution modeling.
© 2024 The Author(s). Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures








References
-
- Abdi, A. (2013). Integrating open access geospatial data to map the habitat suitability of the declining corn bunting (Miliaria calandra). ISPRS International Journal of Geo‐Information, 2(4), 935–954. 10.3390/ijgi2040935 - DOI
-
- Aiello‐Lammens, M. E. , Boria, R. A. , Radosavljevic, A. , Vilela, B. , & Anderson, R. P. (2015). spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38(5), 541–545. 10.1111/ecog.01132 - DOI
-
- Allouche, O. , Tsoar, A. , & Kadmon, R. (2006). Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology, 43(6), 1223–1232. 10.1111/j.1365-2664.2006.01214.x - DOI
-
- Andersen, B. R. , McGuire, L. P. , Wigley, T. B. , Miller, D. A. , & Stevens, R. D. (2022). Habitat associations of overwintering bats in managed pine forest landscapes. Forests, 13(5), 803. 10.3390/f13050803 - DOI
LinkOut - more resources
Full Text Sources

