Single-cell profiling indicates a high similarity between immune cells in the cerebrospinal fluid and in meningeal ectopic lymphoid tissue in experimental autoimmune encephalomyelitis
- PMID: 38933267
- PMCID: PMC11199773
- DOI: 10.3389/fimmu.2024.1400641
Single-cell profiling indicates a high similarity between immune cells in the cerebrospinal fluid and in meningeal ectopic lymphoid tissue in experimental autoimmune encephalomyelitis
Abstract
Background and objectives: B cell depleting anti-CD20 monoclonal antibodies (aCD20 mAbs) are highly effective in treatment of multiple sclerosis (MS) but fail to halt the formation of meningeal ectopic lymphoid tissue (mELT) in the murine model experimental autoimmune encephalomyelitis (EAE). While mELT can be examined in EAE, it is not accessible in vivo in MS patients. Our key objectives were to compare the immune cells in cerebrospinal fluid (CSF), which is accessible in patients, with those in mELT, and to study the effects of aCD20 mAbs on CSF and mELT in EAE.
Methods: Applying single cell RNA sequencing, we compared gene expression profiles in immune cells from (1) CSF with mELT and (2) aCD20 mAbs treated with control treated mice in a spontaneous 2D2xTh EAE model.
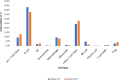
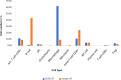
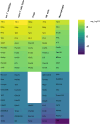
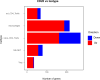
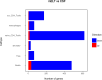
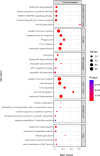
Results: The immune cell composition in CSF and mELT was very similar. Gene expression profiles and pathway enrichment analysis revealed no striking differences between the two compartments. aCD20 mAbs led not only to a virtually complete depletion of B cells in the CSF but also to a reduction of naïve CD4+ T cells and marked increase of macrophages. No remarkable differences in regulated genes or pathways were observed.
Discussion: Our results suggest that immune cells in the CSF may serve as a surrogate for mELT in EAE. Future studies are required to confirm this in MS patients. The observed increase of macrophages in B cell depleted CSF is a novel finding and requires verification in CSF of aCD20 mAbs treated MS patients. Due to unresolved technical challenges, we were unable to study the effects of aCD20 mAbs on mELT. This should be addressed in future studies.
Keywords: B cells; anti-CD20 treatment; ectopic lymphoid tissue; experimental autoimmune encephalomyelitis; multiple sclerosis; single-cell sequencing.
Copyright © 2024 Georgieva, Diddens, Friedrich, Lepennetier, Brand and Lehmann-Horn.
Conflict of interest statement
KL-H has received research support to TUM from Novartis and honoraria and compensation for travel expenses from Novartis, FHLR, Biogen, Teva, Hexal, Bayer, and Merck Serono. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures




References
-
- Kappos L, Wolinsky JS, Giovannoni G, Arnold DL, Wang Q, Bernasconi C, et al. . Contribution of relapse-independent progression vs relapse-associated worsening to overall confirmed disability accumulation in typical relapsing multiple sclerosis in a pooled analysis of 2 randomized clinical trials. JAMA Neurol. (2020) 77:1132–40. doi: 10.1001/jamaneurol.2020.1568 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

