Spatial transcriptomics combined with single-nucleus RNA sequencing reveals glial cell heterogeneity in the human spinal cord
- PMID: 38934400
- PMCID: PMC11881709
- DOI: 10.4103/NRR.NRR-D-23-01876
Spatial transcriptomics combined with single-nucleus RNA sequencing reveals glial cell heterogeneity in the human spinal cord
Abstract
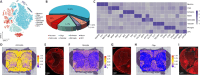
JOURNAL/nrgr/04.03/01300535-202511000-00032/figure1/v/2024-12-20T164640Z/r/image-tiff Glial cells play crucial roles in regulating physiological and pathological functions, including sensation, the response to infection and acute injury, and chronic neurodegenerative disorders. Glial cells include astrocytes, microglia, and oligodendrocytes in the central nervous system, and satellite glial cells and Schwann cells in the peripheral nervous system. Despite the greater understanding of glial cell types and functional heterogeneity achieved through single-cell and single-nucleus RNA sequencing in animal models, few studies have investigated the transcriptomic profiles of glial cells in the human spinal cord. Here, we used high-throughput single-nucleus RNA sequencing and spatial transcriptomics to map the cellular and molecular heterogeneity of astrocytes, microglia, and oligodendrocytes in the human spinal cord. To explore the conservation and divergence across species, we compared these findings with those from mice. In the human spinal cord, astrocytes, microglia, and oligodendrocytes were each divided into six distinct transcriptomic subclusters. In the mouse spinal cord, astrocytes, microglia, and oligodendrocytes were divided into five, four, and five distinct transcriptomic subclusters, respectively. The comparative results revealed substantial heterogeneity in all glial cell types between humans and mice. Additionally, we detected sex differences in gene expression in human spinal cord glial cells. Specifically, in all astrocyte subtypes, the levels of NEAT1 and CHI3L1 were higher in males than in females, whereas the levels of CST3 were lower in males than in females. In all microglial subtypes, all differentially expressed genes were located on the sex chromosomes. In addition to sex-specific gene differences, the levels of MT-ND4 , MT2A , MT-ATP6 , MT-CO3 , MT-ND2 , MT-ND3 , and MT-CO2 in all spinal cord oligodendrocyte subtypes were higher in females than in males. Collectively, the present dataset extensively characterizes glial cell heterogeneity and offers a valuable resource for exploring the cellular basis of spinal cord-related illnesses, including chronic pain, amyotrophic lateral sclerosis, and multiple sclerosis.
Copyright © 2025 Neural Regeneration Research.
Conflict of interest statement
Figures






Similar articles
-
Heterogeneity and Molecular Markers for CNS Glial Cells Revealed by Single-Cell Transcriptomics.Cell Mol Neurobiol. 2022 Nov;42(8):2629-2642. doi: 10.1007/s10571-021-01159-3. Epub 2021 Oct 26. Cell Mol Neurobiol. 2022. PMID: 34704168 Free PMC article. Review.
-
Induction of metallothionein in astrocytes and microglia in the spinal cord from the myelin-deficient jimpy mouse.Brain Res. 1997 Sep 5;767(2):345-55. doi: 10.1016/s0006-8993(97)00628-8. Brain Res. 1997. PMID: 9367267
-
Single-cell sequencing reveals glial cell involvement in development of neuropathic pain via myelin sheath lesion formation in the spinal cord.J Neuroinflammation. 2024 Aug 31;21(1):213. doi: 10.1186/s12974-024-03207-3. J Neuroinflammation. 2024. PMID: 39217340 Free PMC article.
-
Spatial transcriptomics and single-nucleus RNA sequencing reveal a transcriptomic atlas of adult human spinal cord.Elife. 2024 Jan 30;12:RP92046. doi: 10.7554/eLife.92046. Elife. 2024. PMID: 38289829 Free PMC article.
-
Diversity and Function of Glial Cell Types in Multiple Sclerosis.Trends Immunol. 2021 Mar;42(3):228-247. doi: 10.1016/j.it.2021.01.005. Epub 2021 Feb 13. Trends Immunol. 2021. PMID: 33593693 Free PMC article. Review.
Cited by
-
Sex-Specific Properties of Astrocytes: From Development to Evolutionary Insights.Neurochem Res. 2025 Aug 13;50(4):267. doi: 10.1007/s11064-025-04512-w. Neurochem Res. 2025. PMID: 40802016 Free PMC article. Review.
-
Sex and age differences in glia and myelin in nonhuman primate and human spinal cords: implications for pathology.Cell Death Discov. 2025 Apr 2;11(1):129. doi: 10.1038/s41420-025-02425-9. Cell Death Discov. 2025. PMID: 40175332 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

