P53 Activation Promotes Maturational Characteristics of Pluripotent Stem Cell-Derived Cardiomyocytes in 3-Dimensional Suspension Culture Via FOXO-FOXM1 Regulation
- PMID: 38934864
- PMCID: PMC11255683
- DOI: 10.1161/JAHA.123.033155
P53 Activation Promotes Maturational Characteristics of Pluripotent Stem Cell-Derived Cardiomyocytes in 3-Dimensional Suspension Culture Via FOXO-FOXM1 Regulation
Abstract
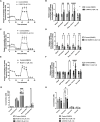
Background: Current protocols generate highly pure human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) in vitro that recapitulate characteristics of mature in vivo cardiomyocytes. Yet, a risk of arrhythmias exists when hiPSC-CMs are injected into large animal models. Thus, understanding hiPSC-CM maturational mechanisms is crucial for clinical translation. Forkhead box (FOX) transcription factors regulate postnatal cardiomyocyte maturation through a balance between FOXO and FOXM1. We also previously demonstrated that p53 activation enhances hiPSC-CM maturation. Here, we investigate whether p53 activation modulates the FOXO/FOXM1 balance to promote hiPSC-CM maturation in 3-dimensional suspension culture.
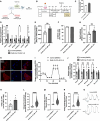
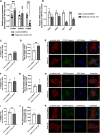
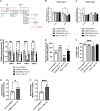
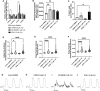
Methods and results: Three-dimensional cultures of hiPSC-CMs were treated with Nutlin-3a (p53 activator, 10 μM), LOM612 (FOXO relocator, 5 μM), AS1842856 (FOXO inhibitor, 1 μM), or RCM-1 (FOXM1 inhibitor, 1 μM), starting 2 days after onset of beating, with dimethyl sulfoxide (0.2% vehicle) as control. P53 activation promoted hiPSC-CM metabolic and electrophysiological maturation alongside FOXO upregulation and FOXM1 downregulation, in n=3 to 6 per group for all assays. FOXO inhibition significantly decreased expression of cardiac-specific markers such as TNNT2. In contrast, FOXO activation or FOXM1 inhibition promoted maturational characteristics such as increased contractility, oxygen consumption, and voltage peak maximum upstroke velocity, in n=3 to 6 per group for all assays. Further, by single-cell RNA sequencing of n=2 LOM612-treated cells compared with dimethyl sulfoxide, LOM612-mediated FOXO activation promoted expression of cardiac maturational pathways.
Conclusions: We show that p53 activation promotes FOXO and suppresses FOXM1 during 3-dimensional hiPSC-CM maturation. These results expand our understanding of hiPSC-CM maturational mechanisms in a clinically-relevant 3-dimensional culture system.
Keywords: FOXM1; FOXO; cardiomyocytes; maturation; p53; stem cells.
Figures


References
-
- Hu D, Linders A, Yamak A, Correia C, Kijlstra JD, Garakani A, Xiao L, Milan DJ, van der Meer P, Serra M, et al. Metabolic maturation of human pluripotent stem cell‐derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ Res. 2018;123:1066–1079. doi: 10.1161/CIRCRESAHA.118.313249 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

