PICALM Regulating the Generation of Amyloid β-Peptide to Promote Anthracycline-Induced Cardiotoxicity
- PMID: 38935046
- PMCID: PMC11348153
- DOI: 10.1002/advs.202401945
PICALM Regulating the Generation of Amyloid β-Peptide to Promote Anthracycline-Induced Cardiotoxicity
Abstract
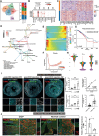
Anthracyclines are chemotherapeutic drugs used to treat solid and hematologic malignancies. However, life-threatening cardiotoxicity, with cardiac dilation and heart failure, is a drawback. A combination of in vivo for single cell/nucleus RNA sequencing and in vitro approaches is used to elucidate the underlying mechanism. Genetic depletion and pharmacological blocking peptides on phosphatidylinositol binding clathrin assembly (PICALM) are used to evaluate the role of PICALM in doxorubicin-induced cardiotoxicity in vivo. Human heart tissue samples are used for verification. Patients with end-stage heart failure and chemotherapy-induced cardiotoxicity have thinner cell membranes compared to healthy controls do. Using the doxorubicin-induced cardiotoxicity mice model, it is possible to replicate the corresponding phenotype in patients. Cellular changes in doxorubicin-induced cardiotoxicity in mice, especially in cardiomyocytes, are identified using single cell/nucleus RNA sequencing. Picalm expression is upregulated only in cardiomyocytes with doxorubicin-induced cardiotoxicity. Amyloid β-peptide production is also increased after doxorubicin treatment, which leads to a greater increase in the membrane permeability of cardiomyocytes. Genetic depletion and pharmacological blocking peptides on Picalm reduce the generation of amyloid β-peptide. This alleviates the doxorubicin-induced cardiotoxicity in vitro and in vivo. In human heart tissue samples of patients with chemotherapy-induced cardiotoxicity, PICALM, and amyloid β-peptide are elevated as well.
Keywords: amyloid β‐peptide; cardiomyocytes; cardiotoxicity.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Cardinale D., Colombo A., Bacchiani G., Tedeschi I., Meroni C. A., Veglia F., Civelli M., Lamantia G., Colombo N., Curigliano G., Fiorentini C., Cipolla C. M., Circulation 2015, 131, 1981. - PubMed
-
- Galan‐Arriola C., Villena‐Gutierrez R., Higuero‐Verdejo M. I., Diaz‐Rengifo I. A., Pizarro G., Lopez G. J., Molina‐Iracheta A., Perez‐Martinez C., Garcia R. D., Gonzalez‐Calle D., Lobo M., Sanchez P. L., Oliver E., Cordoba R., Fuster V., Sanchez‐Gonzalez J., Ibanez B., Cardiovasc. Res. 2021, 117, 1132. - PMC - PubMed
-
- Wu L., Wang L., Du Y., Zhang Y., Ren J., Trends Pharmacol. Sci. 2022, 44, 34. - PubMed
-
- Arinno A., Maneechote C., Khuanjing T., Ongnok B., Prathumsap N., Chunchai T., Arunsak B., Kerdphoo S., Shinlapawittayatorn K., Chattipakorn S. C., Chattipakorn N., Biochem. Pharmacol. 2021, 192, 114743. - PubMed
MeSH terms
Substances
Grants and funding
- 82125004/National Science Fund for Distinguished Young Scholars of China
- JCYJ20220818103414030/Shenzhen Science and Technology Innovation Program
- 82300397/National Natural Science Foundation of China
- 81900335/National Natural Science Foundation of China
- 22275124/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
