Prioritizing drug targets by perturbing biological network response functions
- PMID: 38935814
- PMCID: PMC11236158
- DOI: 10.1371/journal.pcbi.1012195
Prioritizing drug targets by perturbing biological network response functions
Abstract
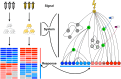
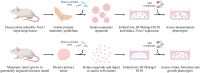
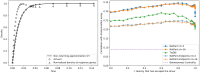
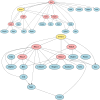
Therapeutic interventions are designed to perturb the function of a biological system. However, there are many types of proteins that cannot be targeted with conventional small molecule drugs. Accordingly, many identified gene-regulatory drivers and downstream effectors are currently undruggable. Drivers and effectors are often connected by druggable signaling and regulatory intermediates. Methods to identify druggable intermediates therefore have general value in expanding the set of targets available for hypothesis-driven validation. Here we identify and prioritize potential druggable intermediates by developing a network perturbation theory, termed NetPert, for response functions of biological networks. Dynamics are defined by a network structure in which vertices represent genes and proteins, and edges represent gene-regulatory interactions and protein-protein interactions. Perturbation theory for network dynamics prioritizes targets that interfere with signaling from driver to response genes. Applications to organoid models for metastatic breast cancer demonstrate the ability of this mathematical framework to identify and prioritize druggable intermediates. While the short-time limit of the perturbation theory resembles betweenness centrality, NetPert is superior in generating target rankings that correlate with previous wet-lab assays and are more robust to incomplete or noisy network data. NetPert also performs better than a related graph diffusion approach. Wet-lab assays demonstrate that drugs for targets identified by NetPert, including targets that are not themselves differentially expressed, are active in suppressing additional metastatic phenotypes.
Copyright: © 2024 Perrone et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: JSB is a founder of and advisor to Neochromosome, Inc., and its parent company Opentrons Labworks, Inc. JSB is an advisor to Dextera Biosciences, Inc. AJE has unlicensed patents related to the use of K14 as a biomarker in breast cancer, US20140336282A1, and for the use of antibody therapeutics in cancer, US2018104331A1. AJE is a consultant for BioNTech. AJE’s spouse is an employee of Immunocore.
Figures

References
-
- Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, et al.. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proceedings of the National Academy of Sciences. 2016;113(7):E854–E863. doi: 10.1073/pnas.1508541113 - DOI - PMC - PubMed
-
- Ewald AJ, Huebner RJ, Palsdottir H, Lee JK, Perez MJ, Jorgens DM, et al.. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. Journal of Cell Science. 2012;125(11):2638–2654. doi: 10.1242/jcs.096875 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

