PCAF promotes R-loop resolution via histone acetylation
- PMID: 38936834
- PMCID: PMC11347145
- DOI: 10.1093/nar/gkae558
PCAF promotes R-loop resolution via histone acetylation
Abstract
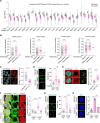
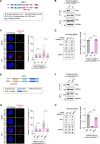
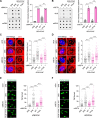
R-loops cause genome instability, disrupting normal cellular functions. Histone acetylation, particularly by p300/CBP-associated factor (PCAF), is essential for maintaining genome stability and regulating cellular processes. Understanding how R-loop formation and resolution are regulated is important because dysregulation of these processes can lead to multiple diseases, including cancer. This study explores the role of PCAF in maintaining genome stability, specifically for R-loop resolution. We found that PCAF depletion promotes the generation of R-loop structures, especially during ongoing transcription, thereby compromising genome stability. Mechanistically, we found that PCAF facilitates histone H4K8 acetylation, leading to recruitment of the a double-strand break repair protein (MRE11) and exonuclease 1 (EXO1) to R-loop sites. These in turn recruit Fanconi anemia (FA) proteins, including FANCM and BLM, to resolve the R-loop structure. Our findings suggest that PCAF, histone acetylation, and FA proteins collaborate to resolve R-loops and ensure genome stability. This study therefore provides novel mechanistic insights into the dynamics of R-loops as well as the role of PCAF in preserving genome stability. These results may help develop therapeutic strategies to target diseases associated with genome instability.
Plain language summary
R-loops are harmful DNA-RNA hybrid structures that cause genome instability, disrupting normal cell functions. This study explored the role of the protein PCAF in resolving R-loops to maintain genome stability. The researchers found that depleting PCAF leads to increased R-loop formation, especially during transcription, compromising the genome. Mechanistically, PCAF facilitates histone acetylation, recruiting proteins like MRE11, EXO1, FANCM and BLM to R-loop sites. These proteins collaborate to resolve R-loop structures. The findings suggest that PCAF, histone acetylation, and these repair proteins work together to untangle R-loops and preserve genome integrity. Understanding this process provides insights into R-loop dynamics and PCAF's role in genome maintenance, potentially leading to therapeutic strategies for diseases associated with genome instability, such as cancer.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Garcia-Muse T., Aguilera A.. R loops: from physiological to pathological Roles. Cell. 2019; 179:604–618. - PubMed
-
- Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. - PubMed
-
- Marnef A., Legube G.. R-loops as Janus-faced modulators of DNA repair. Nat. Cell Biol. 2021; 23:305–313. - PubMed
-
- Petermann E., Lan L., Zou L.. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat. Rev. Mol. Cell Biol. 2022; 23:521–540. - PubMed
-
- Wells J.P., White J., Stirling P.C.. Loops and their composite cancer connections. Trends Cancer. 2019; 5:619–631. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

