A Cullin 5-based complex serves as an essential modulator of ORF9b stability in SARS-CoV-2 replication
- PMID: 38937432
- PMCID: PMC11211426
- DOI: 10.1038/s41392-024-01874-5
A Cullin 5-based complex serves as an essential modulator of ORF9b stability in SARS-CoV-2 replication
Abstract
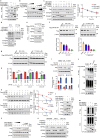
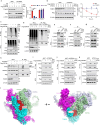
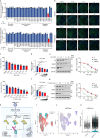
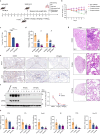
The ORF9b protein, derived from the nucleocapsid's open-reading frame in both SARS-CoV and SARS-CoV-2, serves as an accessory protein crucial for viral immune evasion by inhibiting the innate immune response. Despite its significance, the precise regulatory mechanisms underlying its function remain elusive. In the present study, we unveil that the ORF9b protein of SARS-CoV-2, including emerging mutant strains like Delta and Omicron, can undergo ubiquitination at the K67 site and subsequent degradation via the proteasome pathway, despite certain mutations present among these strains. Moreover, our investigation further uncovers the pivotal role of the translocase of the outer mitochondrial membrane 70 (TOM70) as a substrate receptor, bridging ORF9b with heat shock protein 90 alpha (HSP90α) and Cullin 5 (CUL5) to form a complex. Within this complex, CUL5 triggers the ubiquitination and degradation of ORF9b, acting as a host antiviral factor, while HSP90α functions to stabilize it. Notably, treatment with HSP90 inhibitors such as GA or 17-AAG accelerates the degradation of ORF9b, leading to a pronounced inhibition of SARS-CoV-2 replication. Single-cell sequencing data revealed an up-regulation of HSP90α in lung epithelial cells from COVID-19 patients, suggesting a potential mechanism by which SARS-CoV-2 may exploit HSP90α to evade the host immunity. Our study identifies the CUL5-TOM70-HSP90α complex as a critical regulator of ORF9b protein stability, shedding light on the intricate host-virus immune response dynamics and offering promising avenues for drug development against SARS-CoV-2 in clinical settings.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

