Design of a covalent protein-protein interaction inhibitor of SRPKs to suppress angiogenesis and invasion of cancer cells
- PMID: 38937565
- PMCID: PMC11211491
- DOI: 10.1038/s42004-024-01230-2
Design of a covalent protein-protein interaction inhibitor of SRPKs to suppress angiogenesis and invasion of cancer cells
Abstract
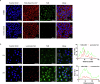
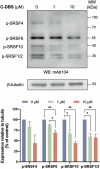
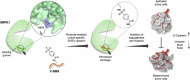
Serine-arginine (SR) proteins are splicing factors that play essential roles in both constitutive and alternative pre-mRNA splicing. Phosphorylation of their C-terminal RS domains by SR protein kinases (SRPKs) regulates their localization and diverse cellular activities. Dysregulation of phosphorylation has been implicated in many human diseases, including cancers. Here, we report the development of a covalent protein-protein interaction inhibitor, C-DBS, that targets a lysine residue within the SRPK-specific docking groove to block the interaction and phosphorylation of the prototypic SR protein SRSF1. C-DBS exhibits high specificity and conjugation efficiency both in vitro and in cellulo. This self-cell-penetrating inhibitor attenuates the phosphorylation of endogenous SR proteins and subsequently inhibits the angiogenesis, migration, and invasion of cancer cells. These findings provide a new foundation for the development of covalent SRPK inhibitors for combatting diseases such as cancer and viral infections and overcoming the resistance encountered by ATP-competitive inhibitors.
© 2024. The Author(s).
Conflict of interest statement
Gongli Cai, Yishu Bao, Qingyun Li, Jiang Xia, and Jacky Chi Ki Ngo are co-applicants for a provisional patent based on this work. All other authors declare no competing interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

