T cell-mediated microglial activation triggers myelin pathology in a mouse model of amyloidosis
- PMID: 38937583
- PMCID: PMC11303250
- DOI: 10.1038/s41593-024-01682-8
T cell-mediated microglial activation triggers myelin pathology in a mouse model of amyloidosis
Abstract
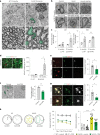
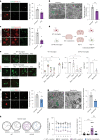
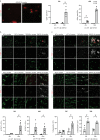
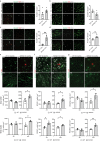
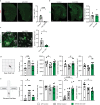
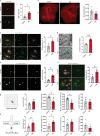
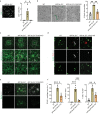
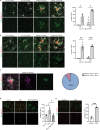
Age-related myelin damage induces inflammatory responses, yet its involvement in Alzheimer's disease remains uncertain, despite age being a major risk factor. Using a mouse model of Alzheimer's disease, we found that amyloidosis itself triggers age-related oligodendrocyte and myelin damage. Mechanistically, CD8+ T cells promote the progressive accumulation of abnormally interferon-activated microglia that display myelin-damaging activity. Thus, our data suggest that immune responses against myelinating oligodendrocytes may contribute to neurodegenerative diseases with amyloidosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

