trans-Interacting Plasma Membrane Proteins and Binding Partner Identification
- PMID: 38937710
- PMCID: PMC11533685
- DOI: 10.1021/acs.jproteome.4c00289
trans-Interacting Plasma Membrane Proteins and Binding Partner Identification
Abstract
Plasma membrane proteins (PMPs) play critical roles in a myriad of physiological and disease conditions. A unique subset of PMPs functions through interacting with each other in trans at the interface between two contacting cells. These trans-interacting PMPs (tiPMPs) include adhesion molecules and ligands/receptors that facilitate cell-cell contact and direct communication between cells. Among the tiPMPs, a significant number have apparent extracellular binding domains but remain orphans with no known binding partners. Identification of their potential binding partners is therefore important for the understanding of processes such as organismal development and immune cell activation. While a number of methods have been developed for the identification of protein binding partners in general, very few are applicable to tiPMPs, which interact in a two-dimensional fashion with low intrinsic binding affinities. In this review, we present the significance of tiPMP interactions, the challenges of identifying binding partners for tiPMPs, and the landscape of method development. We describe current avidity-based screening approaches for identifying novel tiPMP binding partners and discuss their advantages and limitations. We conclude by highlighting the importance of developing novel methods of identifying new tiPMP interactions for deciphering the complex protein interactome and developing targeted therapeutics for diseases.
Keywords: ELISA; Plasma membrane proteins; avidity-enhanced screening; protein interactions; protein interactome; pull-down.
Conflict of interest statement
Competing interests
The author has no relevant financial or non-financial interests to disclose.
Figures
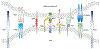
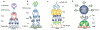
Similar articles
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Topical antibiotics with steroids for chronic suppurative otitis media.Cochrane Database Syst Rev. 2025 Jun 9;6(6):CD013054. doi: 10.1002/14651858.CD013054.pub3. Cochrane Database Syst Rev. 2025. PMID: 40484406
References
-
- Bausch-Fluck D, Hofmann A, Bock T, Frei AP, Cerciello F, Jacobs A, Moest H, Omasits U, Gundry RL, Yoon C, Schiess R, Schmidt A, Mirkowska P, Härtlová A, van Eyk JE, Bourquin JP, Aebersold R, Boheler KR, Zandstra P, & Wollscheid B (2015). A mass spectrometric-derived cell surface protein atlas. PLoS ONE, 10(4). 10.1371/journal.pone.0121314 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

