An aptamer-mediated base editing platform for simultaneous knockin and multiple gene knockout for allogeneic CAR-T cells generation
- PMID: 38937969
- PMCID: PMC11405993
- DOI: 10.1016/j.ymthe.2024.06.033
An aptamer-mediated base editing platform for simultaneous knockin and multiple gene knockout for allogeneic CAR-T cells generation
Abstract
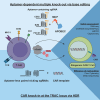
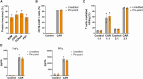
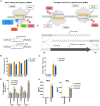
Gene editing technologies hold promise for enabling the next generation of adoptive cellular therapies. In conventional gene editing platforms that rely on nuclease activity, such as clustered regularly interspaced short palindromic repeats CRISPR-associated protein 9 (CRISPR-Cas9), allow efficient introduction of genetic modifications; however, these modifications occur via the generation of DNA double-strand breaks (DSBs) and can lead to unwanted genomic alterations and genotoxicity. Here, we apply a novel modular RNA aptamer-mediated Pin-point base editing platform to simultaneously introduce multiple gene knockouts and site-specific integration of a transgene in human primary T cells. We demonstrate high editing efficiency and purity at all target sites and significantly reduced frequency of chromosomal translocations compared with the conventional CRISPR-Cas9 system. Site-specific knockin of a chimeric antigen receptor and multiplex gene knockout are achieved within a single intervention and without the requirement for additional sequence-targeting components. The ability to perform complex genome editing efficiently and precisely highlights the potential of the Pin-point platform for application in a range of advanced cell therapies.
Keywords: CRISPR; advanced genome editing; allogeneic cell therapy; base editing; gene editing; knockin; knockout; multiple base editing; multiple gene knockout; transgene integration.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M. Francescatto, M. Firth, J.L.T., D.P., J. Sumner, and B.T. are all current or past (while engaged in the research project) employees of AstraZeneca. I.P., R.B., J.H., B.J., O.M., J. Stombaugh, K.H., T.S., Z.S., C.W., A.v.B.S., and J.J.L. are current or past (while engaged in the research project) employees at Revvity. Revvity has an exclusive license from Rutgers University to certain base editing patents. Rutgers University and Horizon Discovery Limited have filed patent applications on this work.
Figures





References
-
- Ottaviano G., Georgiadis C., Gkazi S.A., Syed F., Zhan H., Etuk A., Preece R., Chu J., Kubat A., Adams S., et al. Phase 1 clinical trial of CRISPR-engineered CAR19 universal T cells for treatment of children with refractory B cell leukemia. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abq3010. - DOI - PubMed
-
- Benjamin R., Graham C., Yallop D., Jozwik A., Mirci-Danicar O.C., Lucchini G., Pinner D., Jain N., Kantarjian H., Boissel N., et al. Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: results of two phase 1 studies. The Lancet. 2020;396:1885–1894. doi: 10.1016/S0140-6736(20)32334-5. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

