Breed-Driven Microbiome Heterogeneity Regulates Intestinal Stem Cell Proliferation via Lactobacillus-Lactate-GPR81 Signaling
- PMID: 38937989
- PMCID: PMC11434115
- DOI: 10.1002/advs.202400058
Breed-Driven Microbiome Heterogeneity Regulates Intestinal Stem Cell Proliferation via Lactobacillus-Lactate-GPR81 Signaling
Abstract
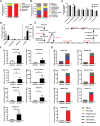
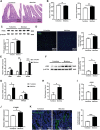
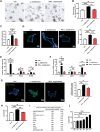
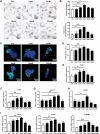
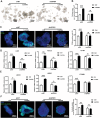
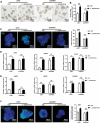
Genetically lean and obese individuals have distinct intestinal microbiota and function. However, the underlying mechanisms of the microbiome heterogeneity and its regulation on epithelial function such as intestinal stem cell (ISC) fate remain unclear. Employing pigs of genetically distinct breeds (obese Meishan and lean Yorkshire), this study reveals transcriptome-wide variations in microbial ecology of the jejunum, characterized by enrichment of active Lactobacillus species, notably the predominant Lactobacillus amylovorus (L. amylovorus), and lactate metabolism network in obese breeds. The L. amylovorus-dominant heterogeneity is paralleled with epithelial functionality difference as reflected by highly expressed GPR81, more proliferative ISCs and activated Wnt/β-catenin signaling. Experiments using in-house developed porcine jejunal organoids prove that live L. amylovorus and its metabolite lactate promote intestinal organoid growth. Mechanistically, L. amylovorus and lactate activate Wnt/β-catenin signaling in a GPR81-dependent manner to promote ISC-mediated epithelial proliferation. However, heat-killed L. amylovorus fail to cause these changes. These findings uncover a previously underrepresented role of L. amylovorus in regulating jejunal stem cells via Lactobacillus-lactate-GPR81 axis, a key mechanism bridging breed-driven intestinal microbiome heterogeneity with ISC fate. Thus, results from this study provide new insights into the role of gut microbiome and stem cell interactions in maintaining intestinal homeostasis.
Keywords: intestinal stem cell; lactobacillus; microbiome; small intestine; wnt/β‐catenin signaling.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
References
-
- Gehart H., Clevers H., Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
