Unexpected complexity in the molecular diagnosis of spastic paraplegia 11
- PMID: 38938072
- PMCID: PMC11211614
- DOI: 10.1002/mgg3.2475
Unexpected complexity in the molecular diagnosis of spastic paraplegia 11
Abstract
Background: Spastic paraplegia 11 (SPG11) is the most prevalent form of autosomal recessive hereditary spastic paraplegia, resulting from biallelic pathogenic variants in the SPG11 gene (MIM *610844).
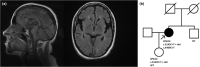
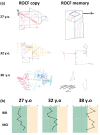
Methods: The proband is a 36-year-old female referred for genetic evaluation due to cognitive dysfunction, gait impairment, and corpus callosum atrophy (brain MRI was normal at 25-years-old). Diagnostic approaches included CGH array, next-generation sequencing, and whole transcriptome sequencing.
Results: CGH array revealed a 180 kb deletion located upstream of SPG11. Sequencing of SPG11 uncovered two rare single nucleotide variants: the novel variant c.3143C>T in exon 17 (in cis with the deletion), and the previously reported pathogenic variant c.6409C>T in exon 34 (in trans). Whole transcriptome sequencing revealed that the variant c.3143C>T caused exon 17 skipping.
Conclusion: We report a novel sequence variant in the SPG11 gene resulting in exon 17 skipping, which, along with a nonsense variant, causes Spastic Paraplegia 11 in our proband. In addition, a deletion upstream of SPG11 was identified in the patient, whose implication in the phenotype remains uncertain. Nonetheless, the deletion apparently affects cis-regulatory elements of the gene, suggesting a potential new pathogenic mechanism underlying the disease in a subset of undiagnosed patients. Our findings further support the hypothesis that the origin of thin corpus callosum in patients with SPG11 is of progressive nature.
Keywords: SPG11; cis‐regulatory elements; genetic diagnosis; spastic paraplegia 11; whole transcriptome sequencing.
© 2024 The Author(s). Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
There are no conflicts of interest associated with this publication.
Figures


References
-
- Bauer, P. , Winner, B. , Schüle, R. , Bauer, C. , Häfele, V. , Hehr, U. , Bonin, M. , Walter, M. , Karle, K. , Ringer, T. M. , Rieß, O. , Winkler, J. , & Schöls, L. (2009). Identification of a heterozygous genomic deletion in the spatacsin gene in SPG11 patients using high‐resolution comparative genomic hybridization. Neurogenetics, 10(1), 43–48. - PubMed
-
- Benito‐Sanz, S. , Aza‐Carmona, M. , Rodríguez‐Estevez, A. , Rica‐Etxebarria, I. , Gracia, R. , Campos‐Barros, Á. , & Heath, K. E. (2012). Identification of the first PAR1 deletion encompassing upstream SHOX enhancers in a family with idiopathic short stature. European Journal of Human Genetics, 20(1), 125–127. - PMC - PubMed
-
- Boutry, M. , Branchu, J. , Lustremant, C. , Pujol, C. , Pernelle, J. , Matusiak, R. , Seyer, A. , Poirel, M. , Chu‐van, E. , Pierga, A. , Dobrenis, K. , Puech, J. P. , Caillaud, C. , Durr, A. , Brice, A. , Colsch, B. , Mochel, F. , el Hachimi, K. H. , Stevanin, G. , & Darios, F. (2018). Inhibition of lysosome membrane recycling causes accumulation of gangliosides that contribute to neurodegeneration. Cell Reports, 23(13), 3813–3826. - PMC - PubMed
-
- Branchu, J. , Boutry, M. , Sourd, L. , Depp, M. , Leone, C. , Corriger, A. , Vallucci, M. , Esteves, T. , Matusiak, R. , Dumont, M. , Muriel, M. P. , Santorelli, F. M. , Brice, A. , el Hachimi, K. H. , Stevanin, G. , & Darios, F. (2017). Loss of spatacsin function alters lysosomal lipid clearance leading to upper and lower motor neuron degeneration. Neurobiology of Disease, 102, 21–37. - PMC - PubMed
-
- Casali, C. , Valente, E. M. , Bertini, E. , Montagna, G. , Criscuolo, C. , De Michele, G. , Villanova, M. , Damiano, M. , Pierallini, A. , Brancati, F. , & Scarano, V. (2004). Clinical and genetic studies in hereditary spastic paraplegia with thin corpus callosum. Neurology, 62(2), 262–268. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

