Genome-wide association study and polygenic score assessment of insulin resistance
- PMID: 38938516
- PMCID: PMC11208314
- DOI: 10.3389/fendo.2024.1384103
Genome-wide association study and polygenic score assessment of insulin resistance
Abstract
Insulin resistance (IR) and beta cell dysfunction are the major drivers of type 2 diabetes (T2D). Genome-Wide Association Studies (GWAS) on IR have been predominantly conducted in European populations, while Middle Eastern populations remain largely underrepresented. We conducted a GWAS on the indices of IR (HOMA2-IR) and beta cell function (HOMA2-%B) in 6,217 non-diabetic individuals from the Qatar Biobank (QBB; Discovery cohort; n = 2170, Replication cohort; n = 4047) with and without body mass index (BMI) adjustment. We also developed polygenic scores (PGS) for HOMA2-IR and compared their performance with a previously derived PGS for HOMA-IR (PGS003470). We replicated 11 loci that have been previously associated with HOMA-IR and 24 loci that have been associated with HOMA-%B, at nominal statistical significance. We also identified a novel locus associated with beta cell function near VEGFC gene, tagged by rs61552983 (P = 4.38 × 10-8). Moreover, our best performing PGS (Q-PGS4; Adj R2 = 0.233 ± 0.014; P = 1.55 x 10-3) performed better than PGS003470 (Adj R2 = 0.194 ± 0.014; P = 5.45 x 10-2) in predicting HOMA2-IR in our dataset. This is the first GWAS on HOMA2 and the first GWAS conducted in the Middle East focusing on IR and beta cell function. Herein, we report a novel locus in VEGFC that is implicated in beta cell dysfunction. Inclusion of under-represented populations in GWAS has potentials to provide important insights into the genetic architecture of IR and beta cell function.
Keywords: GWAS; beta cell; insulin resistance; polygenic score; type 2 diabetes.
Copyright © 2024 Aliyu, Umlai, Toor, Elashi, Al-Sarraj, Abou−Samra, Suhre and Albagha.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
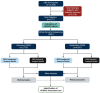
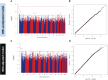
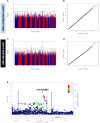
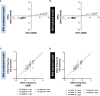

References
-
- Federation ID. IDF diabetes atlas ninth. (2019) 9(1):5–9. Dunia: Idf.
-
- Safiri S, Karamzad N, Kaufman JS, Bell AW, Nejadghaderi SA, Sullman MJM, et al. . Prevalence, deaths and disability-adjusted-life-years (DALYs) due to type 2 diabetes and its attributable risk factors in 204 countries and territories, 1990-2019: results from the global burden of disease study 2019. Front Endocrinol (Lausanne). (2022) 13:838027. doi: 10.3389/fendo.2022.838027 - DOI - PMC - PubMed
-
- Samya Ahmad Al A, Dahlia Mustafa H, Azza Mustafa M, Joelle B. SMART population screening and management in Qatar. Int J Diabetes Clin Res. (2019) 6(1). doi: 10.23937/2377-3634 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

