Respiratory syncytial virus NS1 inhibits anti-viral Interferon-α-induced JAK/STAT signaling, by limiting the nuclear translocation of STAT1
- PMID: 38938568
- PMCID: PMC11208467
- DOI: 10.3389/fimmu.2024.1395809
Respiratory syncytial virus NS1 inhibits anti-viral Interferon-α-induced JAK/STAT signaling, by limiting the nuclear translocation of STAT1
Abstract
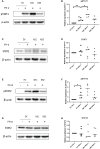
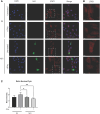
Human respiratory viruses are the most prevalent cause of disease in humans, with the highly infectious RSV being the leading cause of infant bronchiolitis and viral pneumonia. Responses to type I IFNs are the primary defense against viral infection. However, RSV proteins have been shown to antagonize type I IFN-mediated antiviral innate immunity, specifically dampening intracellular IFN signaling. Respiratory epithelial cells are the main target for RSV infection. In this study, we found RSV-NS1 interfered with the IFN-α JAK/STAT signaling pathway of epithelial cells. RSV-NS1 expression significantly enhanced IFN-α-mediated phosphorylation of STAT1, but not pSTAT2; and neither STAT1 nor STAT2 total protein levels were affected by RSV-NS1. However, expression of RSV-NS1 significantly reduced ISRE and GAS promoter activity and anti-viral IRG expression. Further mechanistic studies demonstrated RSV-NS1 bound STAT1, with protein modeling indicating a possible interaction site between STAT1 and RSV-NS1. Nuclear translocation of STAT1 was reduced in the presence of RSV-NS1. Additionally, STAT1's interaction with the nuclear transport adapter protein, KPNA1, was also reduced, suggesting a mechanism by which RSV blocks STAT1 nuclear translocation. Indeed, reducing STAT1's access to the nucleus may explain RSV's suppression of IFN JAK/STAT promoter activation and antiviral gene induction. Taken together these results describe a novel mechanism by which RSV controls antiviral IFN-α JAK/STAT responses, which enhances our understanding of RSV's respiratory disease progression.
Keywords: Interferon; JAK/STAT signaling; RSV; nuclear translocation; viral immune evasion.
Copyright © 2024 Efstathiou, Zhang, Kandwal, Fayne, Molloy and Stevenson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

