Proteome encoded determinants of protein sorting into extracellular vesicles
- PMID: 38938677
- PMCID: PMC11080751
- DOI: 10.1002/jex2.120
Proteome encoded determinants of protein sorting into extracellular vesicles
Abstract
Extracellular vesicles (EVs) are membranous structures released by cells into the extracellular space and are thought to be involved in cell-to-cell communication. While EVs and their cargo are promising biomarker candidates, sorting mechanisms of proteins to EVs remain unclear. In this study, we ask if it is possible to determine EV association based on the protein sequence. Additionally, we ask what the most important determinants are for EV association. We answer these questions with explainable AI models, using human proteome data from EV databases to train and validate the model. It is essential to correct the datasets for contaminants introduced by coarse EV isolation workflows and for experimental bias caused by mass spectrometry. In this study, we show that it is indeed possible to predict EV association from the protein sequence: a simple sequence-based model for predicting EV proteins achieved an area under the curve of 0.77 ± 0.01, which increased further to 0.84 ± 0.00 when incorporating curated post-translational modification (PTM) annotations. Feature analysis shows that EV-associated proteins are stable, polar, and structured with low isoelectric point compared to non-EV proteins. PTM annotations emerged as the most important features for correct classification; specifically, palmitoylation is one of the most prevalent EV sorting mechanisms for unique proteins. Palmitoylation and nitrosylation sites are especially prevalent in EV proteins that are determined by very strict isolation protocols, indicating they could potentially serve as quality control criteria for future studies. This computational study offers an effective sequence-based predictor of EV associated proteins with extensive characterisation of the human EV proteome that can explain for individual proteins which factors contribute to their EV association.
Keywords: biomarkers; extracellular vesicles; human proteome; machine learning; post‐translational modification.
© 2024 The Authors. Journal of Extracellular Biology published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
Research of Katharina Waury, Dea Gogishvili, Charlotte E. Teunissen, and Sanne Abeln are supported by the European Commission Marie Curie International Training Network, grant agreement No 860197, the MIRIADE project. Charlotte E. Teunissen is supported by JPND (bPRIDE), Health Holland, the Dutch Research Council (ZonMW), Alzheimer Drug Discovery Foundation, The Selfridges Group Foundation, Alzheimer Netherlands, Alzheimer Association. Charlotte E. Teunissen is recipient of ABOARD, which is a public‐private partnership receiving funding from ZonMW (#73305095007) and HealthHolland, Topsector Life Sciences & Health (PPP‐allowance; #LSHM20106). More than 30 partners participate in ABOARD. ABOARD also receives funding from Edwin Bouw Fonds and Gieskes‐Strijbisfonds. Charlotte E. Teunissen has a collaboration contract with ADx, Neurosciences, Quanterix and Eli Lilly, performed contract research or received grants from AC‐Immune, Axon Neurosciences, Biogen, Brainstorm Therapeutics, Celgene, EIP Pharma, Eisai, PeopleBio, Roche, Toyama, Vivoryon. She serves on editorial boards of Medidact Neurologie/Springer, Alzheimer Research and Therapy, Neurology: Neuroimmunology & Neuroinflammation, and is editor of a Neuromethods book Springer.
Figures
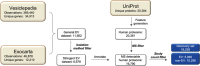

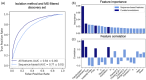


References
-
- Anand, S. , Samuel, M. , Kumar, S. , & Mathivanan, S. (2019). Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochimica Et Biophysica Acta (BBA)‐Proteins and Proteomics, 1867(12), 140203. - PubMed
-
- Ban, J.‐J. , Lee, M. , Im, W. , & Kim, M. (2015). Low pH increases the yield of exosome isolation. Biochemical and Biophysical Research Communications, 461, 76–79. - PubMed
LinkOut - more resources
Full Text Sources
