CRISPR-Cas9 screening reveals a distinct class of MHC-I binders with precise HLA-peptide recognition
- PMID: 38939106
- PMCID: PMC11209011
- DOI: 10.1016/j.isci.2024.110120
CRISPR-Cas9 screening reveals a distinct class of MHC-I binders with precise HLA-peptide recognition
Abstract
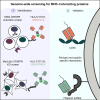
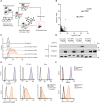
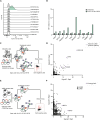
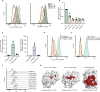
Human leukocyte antigen (HLA) class-I molecules present fragments of the cellular proteome to the T cell receptor (TCR) of cytotoxic T cells to control infectious diseases and cancer. The large number of combinations of HLA class-I allotypes and peptides allows for highly specific and dedicated low-affinity interactions to a diverse array of TCRs and natural killer (NK) cell receptors. Whether the divergent HLA class-I peptide complex is exclusive for interactions with these proteins is unknown. Using genome-wide CRISPR-Cas9 activation and knockout screens, we identified peptide-specific HLA-C∗07 combinations that can interact with the surface molecules CD55 and heparan sulfate. These interactions closely resemble the HLA class-I interaction with the TCR regarding both the affinity range and the specificity of the peptide and HLA allele. These findings indicate that various proteins can specifically bind HLA class-I peptide complexes due to their polymorphic nature, which suggests there are more interactions like the ones we describe here.
Keywords: Biochemistry; Bioinformatics; Biological sciences; Immunology; Natural sciences.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Ekeruche-Makinde J., Miles J.J., van den Berg H.A., Skowera A., Cole D.K., Dolton G., Schauenburg A.J.A., Tan M.P., Pentier J.M., Llewellyn-Lacey S., et al. Peptide length determines the outcome of TCR/peptide-MHCI engagement. Blood. 2013;121:1112–1123. doi: 10.1182/blood-2012-06-437202. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

