Polygenic Risk, Rare Variants, and Family History: Independent and Additive Effects on Coronary Heart Disease
- PMID: 38939477
- PMCID: PMC11198423
- DOI: 10.1016/j.jacadv.2023.100567
Polygenic Risk, Rare Variants, and Family History: Independent and Additive Effects on Coronary Heart Disease
Abstract
Background: Genetic factors are not included in prediction models for coronary heart disease (CHD).
Objectives: The authors assessed the predictive utility of a polygenic risk score (PRS) for CHD (defined as myocardial infarction, coronary revascularization, or cardiovascular death) and whether the risks due to monogenic familial hypercholesterolemia (FH) and family history (FamHx) are independent of and additive to the PRS.
Methods: In UK-biobank participants, PRSCHD was calculated using metaGRS, and 10-year risk for incident CHD was estimated using the pooled cohort equations (PCE). The area under the curve (AUC) of the receiver operator curve and net reclassification improvement (NRI) were assessed. FH was defined as the presence of a pathogenic or likely pathogenic variant in LDLR, APOB, or PCSK9. FamHx was defined as a diagnosis of CHD in first-degree relatives. Independent and additive effects of PRSCHD, FH, and FamHx were evaluated in stratified analyses.
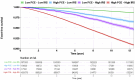
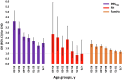
Results: In 323,373 participants with genotype data, the addition of PRSCHD to PCE increased the AUC from 0.759 (95% CI: 0.755-0.763) to 0.773 (95% CI: 0.769-0.777). The AUC and NRIEvent for PRSCHD were higher before the age of 55 years. Of 199,997 participants with exome sequence data, 10,000 had a PRSCHD ≥95th percentile (PRSP95), 673 had FH, and 46,163 had FamHx. The CHD risk associated with PRSP95 was independent of FH and FamHx. The risks associated with combinations of PRSCHD, FH, and FamHx were additive and comprehensive estimates could be obtained by multiplying the risk from each genetic factor.
Conclusions: Incorporating PRSCHD into the PCE improves risk prediction for CHD, especially at younger ages. The associations of PRSCHD, FH, and FamHx with CHD were independent and additive.
Keywords: cardiovascular diseases; coronary disease; genetic predisposition to disease; genome-wide association study; risk assessment.
© 2023 The Authors.
Conflict of interest statement
This study was funded by grants U01HG06379 from the 10.13039/100000051National Human Genome Research Institute and K24HL137010 from the 10.13039/100000050National Heart Lung and Blood Institute. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.PERSPECTIVESCOMPETENCY IN MEDICAL KNOWLEDGE: Incorporating a PRS into the PCE improves CHD risk prediction, particularly in younger individuals and lower-risk individuals (10-year risk <20%). TRANSLATIONAL OUTLOOK: Since the effect of PRS, FH, and family history are independent and additive, these can be used together to obtain a comprehensive assessment of CHD risk.
Figures




References
-
- Arnett D.K., Blumenthal R.S., Albert M.A., et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2019;74(10):e177–e232. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

