TP53 mutations correlate with the non-coding RNA content of small extracellular vesicles in melanoma
- PMID: 38939511
- PMCID: PMC11080853
- DOI: 10.1002/jex2.105
TP53 mutations correlate with the non-coding RNA content of small extracellular vesicles in melanoma
Abstract
Non-coding RNAs (ncRNAs) are important regulators of gene expression. They are expressed not only in cells, but also in cell-derived extracellular vesicles (EVs). The mechanisms controlling their loading and sorting remain poorly understood. Here, we investigated the impact of TP53 mutations on the non-coding RNA content of small melanoma EVs. After purification of small EVs from six different patient-derived melanoma cell lines, we characterized them by small RNA sequencing and lncRNA microarray analysis. We found that TP53 mutations are associated with a specific micro and long non-coding RNA content in small EVs. Then, we showed that long and small non-coding RNAs enriched in TP53 mutant small EVs share a common sequence motif, highly similar to the RNA-binding motif of Sam68, a protein interacting with hnRNP proteins. This protein thus may be an interesting partner of p53, involved in the expression and loading of the ncRNAs. To conclude, our data support the existence of cellular mechanisms associate with TP53 mutations which control the ncRNA content of small EVs in melanoma.
Keywords: Small extracellular vesicle; TP53 mutations; long non‐coding RNA; microRNA.
© 2023 The Authors. Journal of Extracellular Biology published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no competing interests.
Figures
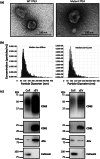
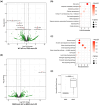
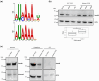

References
-
- Ameli Mojarad, M. , Ameli Mojarad, M. , Shojaee, B. , & Nazemalhosseini‐Mojarad, E. (2022). piRNA: A promising biomarker in early detection of gastrointestinal cancer. Pathology ‐ Research and Practice, 230, 153757. - PubMed
-
- Bielli, P. , Busà, R. , Paronetto, M. P. , & Sette, C. (2011). The RNA‐binding protein Sam68 is a multifunctional player in human cancer. Endocrine‐Related Cancer, 18, R91–102. - PubMed
-
- Cheng, Z. , Lu, C. , Wang, H. , Wang, N. , Cui, S. , Yu, C. , Wang, C. , Zuo, Q. , Wang, S. , Lv, Y. , Yao, M. , Jiang, L. , & Qin, W. (2022). Long noncoding RNA LHFPL3‐AS2 suppresses metastasis of non‐small cell lung cancer by interacting with SFPQ to regulate TXNIP expression. Cancer Letters, 531, 1–13. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
