Sigmoni: classification of nanopore signal with a compressed pangenome index
- PMID: 38940135
- PMCID: PMC11211819
- DOI: 10.1093/bioinformatics/btae213
Sigmoni: classification of nanopore signal with a compressed pangenome index
Abstract
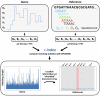
Summary: Improvements in nanopore sequencing necessitate efficient classification methods, including pre-filtering and adaptive sampling algorithms that enrich for reads of interest. Signal-based approaches circumvent the computational bottleneck of basecalling. But past methods for signal-based classification do not scale efficiently to large, repetitive references like pangenomes, limiting their utility to partial references or individual genomes. We introduce Sigmoni: a rapid, multiclass classification method based on the r-index that scales to references of hundreds of Gbps. Sigmoni quantizes nanopore signal into a discrete alphabet of picoamp ranges. It performs rapid, approximate matching using matching statistics, classifying reads based on distributions of picoamp matching statistics and co-linearity statistics, all in linear query time without the need for seed-chain-extend. Sigmoni is 10-100× faster than previous methods for adaptive sampling in host depletion experiments with improved accuracy, and can query reads against large microbial or human pangenomes. Sigmoni is the first signal-based tool to scale to a complete human genome and pangenome while remaining fast enough for adaptive sampling applications.
Availability and implementation: Sigmoni is implemented in Python, and is available open-source at https://github.com/vshiv18/sigmoni.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
S.K. has received travel funding from Oxford Nanopore Technologies Limited.
Figures

Update of
-
Sigmoni: classification of nanopore signal with a compressed pangenome index.bioRxiv [Preprint]. 2023 Aug 30:2023.08.15.553308. doi: 10.1101/2023.08.15.553308. bioRxiv. 2023. Update in: Bioinformatics. 2024 Jun 28;40(Suppl 1):i287-i296. doi: 10.1093/bioinformatics/btae213. PMID: 37645873 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

