RiboDiffusion: tertiary structure-based RNA inverse folding with generative diffusion models
- PMID: 38940178
- PMCID: PMC11211841
- DOI: 10.1093/bioinformatics/btae259
RiboDiffusion: tertiary structure-based RNA inverse folding with generative diffusion models
Abstract
Motivation: RNA design shows growing applications in synthetic biology and therapeutics, driven by the crucial role of RNA in various biological processes. A fundamental challenge is to find functional RNA sequences that satisfy given structural constraints, known as the inverse folding problem. Computational approaches have emerged to address this problem based on secondary structures. However, designing RNA sequences directly from 3D structures is still challenging, due to the scarcity of data, the nonunique structure-sequence mapping, and the flexibility of RNA conformation.
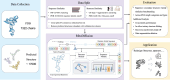
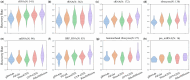
Results: In this study, we propose RiboDiffusion, a generative diffusion model for RNA inverse folding that can learn the conditional distribution of RNA sequences given 3D backbone structures. Our model consists of a graph neural network-based structure module and a Transformer-based sequence module, which iteratively transforms random sequences into desired sequences. By tuning the sampling weight, our model allows for a trade-off between sequence recovery and diversity to explore more candidates. We split test sets based on RNA clustering with different cut-offs for sequence or structure similarity. Our model outperforms baselines in sequence recovery, with an average relative improvement of 11% for sequence similarity splits and 16% for structure similarity splits. Moreover, RiboDiffusion performs consistently well across various RNA length categories and RNA types. We also apply in silico folding to validate whether the generated sequences can fold into the given 3D RNA backbones. Our method could be a powerful tool for RNA design that explores the vast sequence space and finds novel solutions to 3D structural constraints.
Availability and implementation: The source code is available at https://github.com/ml4bio/RiboDiffusion.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


Similar articles
-
R3Design: deep tertiary structure-based RNA sequence design and beyond.Brief Bioinform. 2024 Nov 22;26(1):bbae682. doi: 10.1093/bib/bbae682. Brief Bioinform. 2024. PMID: 39737572 Free PMC article.
-
A Hyperbolic Discrete Diffusion 3D RNA Inverse Folding Model for Functional RNA Design.J Chem Inf Model. 2025 Jul 14;65(13):6568-6584. doi: 10.1021/acs.jcim.5c00527. Epub 2025 Jun 12. J Chem Inf Model. 2025. PMID: 40503717
-
Evolutionary solution for the RNA design problem.Bioinformatics. 2014 May 1;30(9):1250-8. doi: 10.1093/bioinformatics/btu001. Epub 2014 Jan 8. Bioinformatics. 2014. PMID: 24407223
-
Design of RNAs: comparing programs for inverse RNA folding.Brief Bioinform. 2018 Mar 1;19(2):350-358. doi: 10.1093/bib/bbw120. Brief Bioinform. 2018. PMID: 28049135 Free PMC article. Review.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
-
Sifting through the noise: A survey of diffusion probabilistic models and their applications to biomolecules.J Mol Biol. 2025 Mar 15;437(6):168818. doi: 10.1016/j.jmb.2024.168818. Epub 2024 Oct 9. J Mol Biol. 2025. PMID: 39389290 Review.
-
Comprehensive datasets for RNA design, machine learning, and beyond.Sci Rep. 2025 Jul 1;15(1):21417. doi: 10.1038/s41598-025-07041-2. Sci Rep. 2025. PMID: 40594473 Free PMC article.
-
Secondary-Structure-Informed RNA Inverse Design via Relational Graph Neural Networks.Noncoding RNA. 2025 Feb 26;11(2):18. doi: 10.3390/ncrna11020018. Noncoding RNA. 2025. PMID: 40126342 Free PMC article.
-
Computational De Novo Design of Group II Introns Yields Highly Active Ribozymes.Chembiochem. 2025 Jul 18;26(14):e202500356. doi: 10.1002/cbic.202500356. Epub 2025 Jun 30. Chembiochem. 2025. PMID: 40504414 Free PMC article.
-
DRAG: design RNAs as hierarchical graphs with reinforcement learning.Brief Bioinform. 2025 Mar 4;26(2):bbaf106. doi: 10.1093/bib/bbaf106. Brief Bioinform. 2025. PMID: 40079262 Free PMC article.
References
-
- Andronescu M, Fejes AP, Hutter F. et al. A new algorithm for RNA secondary structure design. J Mol Biol 2004;336:607–24. - PubMed
-
- Bank PD. Protein data bank. Nature New Biol 1971;233:223. - PubMed
-
- Benhenda M. ChemGAN challenge for drug discovery: can AI reproduce natural chemical diversity? arXiv, arXiv:1708.08227, 2017, preprint: not peer reviewed.
-
- Busch A, Backofen R.. Info-RNA – a fast approach to inverse RNA folding. Bioinformatics 2006;22:1823–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

