Dispersal history of SARS-CoV-2 in Galicia, Spain
- PMID: 38940448
- PMCID: PMC11742125
- DOI: 10.1002/jmv.29773
Dispersal history of SARS-CoV-2 in Galicia, Spain
Abstract
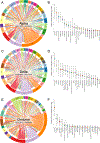
The dynamics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission are influenced by a variety of factors, including social restrictions and the emergence of distinct variants. In this study, we delve into the origins and dissemination of the Alpha, Delta, and Omicron-BA.1 variants of concern in Galicia, northwest Spain. For this, we leveraged genomic data collected by the EPICOVIGAL Consortium and from the GISAID database, along with mobility information from other Spanish regions and foreign countries. Our analysis indicates that initial introductions during the Alpha phase were predominantly from other Spanish regions and France. However, as the pandemic progressed, introductions from Portugal and the United States became increasingly significant. The number of detected introductions varied from 96 and 101 for Alpha and Delta to 39 for Omicron-BA.1. Most of these introductions left a low number of descendants (<10), suggesting a limited impact on the evolution of the pandemic in Galicia. Notably, Galicia's major coastal cities emerged as critical hubs for viral transmission, highlighting their role in sustaining and spreading the virus. This research emphasizes the critical role of regional connectivity in the spread of SARS-CoV-2 and offers essential insights for enhancing public health strategies and surveillance measures.
Keywords: SARS‐CoV‐2; dispersal; phylogeography; variants of concern.
© 2024 The Author(s). Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
Declaration of interest
There are no conflicting interests.
Figures


Update of
-
Dispersal history of SARS-CoV-2 in Galicia, Spain.medRxiv [Preprint]. 2024 Feb 28:2024.02.27.24303385. doi: 10.1101/2024.02.27.24303385. medRxiv. 2024. Update in: J Med Virol. 2024 Jul;96(7):e29773. doi: 10.1002/jmv.29773. PMID: 38463998 Free PMC article. Updated. Preprint.
References
-
- Aksamentov Ivan, Roemer Cornelius, Hodcroft Emma B., and Neher Richard A.. 2021. “Nextclade: Clade Assignment, Mutation Calling and Quality Control for Viral Genomes.” Journal of Open Source Software 6 (67): 3773.
-
- Andrews S 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Ayres Daniel L., Cummings Michael P., Baele Guy, Darling Aaron E., Lewis Paul O., Swofford David L., Huelsenbeck John P., Lemey Philippe, Rambaut Andrew, and Suchard Marc A.. 2019. “BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics.” Systematic Biology 68 (6): 1052–61. - PMC - PubMed
-
- Bager Peter, Wohlfahrt Jan, Bhatt Samir, Stegger Marc, Legarth Rebecca, Møller Camilla Holten, Skov Robert Leo, et al. 2022. “Risk of Hospitalisation Associated with Infection with SARS-CoV-2 Omicron Variant versus Delta Variant in Denmark: An Observational Cohort Study.” The Lancet Infectious Diseases 22 (7): 967–76. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- 06298/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI162611/NH/NIH HHS/United States
- R01 AI153044/AI/NIAID NIH HHS/United States
- EPICOVIGAL FONDO SUPERA-COVID19/Banco Santander-CSIC-CRUE
- G098321N/Research Foundation - Flanders
- Consellería de Sanidade, Xunta de Galicia
- Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia
- U19 AI135995/NH/NIH HHS/United States
- R01 AI162611/AI/NIAID NIH HHS/United States
- F.4515.22/Fonds National de la Recherche Scientifique
- UIDP/50026/2020/Foundation for Science and Technology
- UIDB/50026/2020/Foundation for Science and Technology
- WT_/Wellcome Trust/United Kingdom
- 02/2023-01/2027/DURABLE EU4Health
- U19 AI135995/AI/NIAID NIH HHS/United States
- R01 AI153044/NH/NIH HHS/United States
- C14/18/094/KU Leuven
- 874850/European Union Horizon 2020 project MOOD
- 725422-ReservoirDOCS/European Union's Horizon 2020 Research and Innovation Programme
- 101094685/European Union Horizon RIA 2023 project LEAPS
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

