Characterizing medaka visual features using a high-throughput optomotor response assay
- PMID: 38941325
- PMCID: PMC11213317
- DOI: 10.1371/journal.pone.0302092
Characterizing medaka visual features using a high-throughput optomotor response assay
Abstract
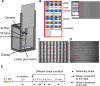
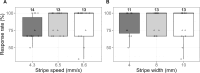
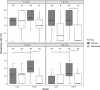
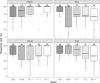
Medaka fish (Oryzias latipes) is a powerful model to study genetics underlying the developmental and functional traits of the vertebrate visual system. We established a simple and high-throughput optomotor response (OMR) assay utilizing medaka larvae to study visual functions including visual acuity and contrast sensitivity. Our assay presents multiple adjustable stripes in motion to individual fish in a linear arena. For that the OMR assay employs a tablet display and the Fish Stripes software to adjust speed, width, color, and contrast of the stripes. Our results demonstrated that optomotor responses were robustly induced by black and white stripes presented from below in the linear-pool-arena. We detected robust strain specific differences in the OMR when comparing long established medaka inbred strains. We observed an interesting training effect upon the initial exposure of larvae to thick stripes, which allowed them to better respond to narrower stripes. The OMR setup and protocol presented here provide an efficient tool for quantitative phenotype mapping, addressing visual acuity, trainability of cortical neurons, color sensitivity, locomotor response, retinal regeneration and others. Our open-source setup presented here provides a crucial prerequisite for ultimately addressing the genetic basis of those processes.
Copyright: © 2024 Suzuki et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

