Quality considerations and major pitfalls for high throughput DNA-based newborn screening for severe combined immunodeficiency and spinal muscular atrophy
- PMID: 38941330
- PMCID: PMC11213327
- DOI: 10.1371/journal.pone.0306329
Quality considerations and major pitfalls for high throughput DNA-based newborn screening for severe combined immunodeficiency and spinal muscular atrophy
Abstract
Background: Many newborn screening programs worldwide have introduced screening for diseases using DNA extracted from dried blood spots (DBS). In Germany, DNA-based assays are currently used to screen for severe combined immunodeficiency (SCID), spinal muscular atrophy (SMA), and sickle cell disease (SCD).
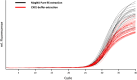
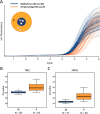
Methods: This study analysed the impact of pre-analytic DNA carry-over in sample preparation on the outcome of DNA-based newborn screening for SCID and SMA and compared the efficacy of rapid extraction versus automated protocols. Additionally, the distribution of T cell receptor excision circles (TREC) on DBS cards, commonly used for routine newborn screening, was determined.
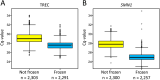
Results: Contaminations from the punching procedure were detected in the SCID and SMA assays in all experimental setups tested. However, a careful evaluation of a cut-off allowed for a clear separation of true positive polymerase chain reaction (PCR) amplifications. Our rapid in-house extraction protocol produced similar amounts compared to automated commercial systems. Therefore, it can be used for reliable DNA-based screening. Additionally, the amount of extracted DNA significantly differs depending on the location of punching within a DBS.
Conclusions: Newborn screening for SMA and SCID can be performed reliably. It is crucial to ensure that affected newborns are not overlooked. Therefore a carefully consideration of potential contaminating factors and the definition of appropriate cut-offs to minimise the risk of false results are of special concern. It is also important to note that the location of punching plays a pivotal role, and therefore an exact quantification of TREC numbers per μl may not be reliable and should therefore be avoided.
Copyright: © 2024 Bzdok et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A Modular Genetic Approach to Newborn Screening from Spinal Muscular Atrophy to Sickle Cell Disease-Results from Six Years of Genetic Newborn Screening.Genes (Basel). 2024 Nov 13;15(11):1467. doi: 10.3390/genes15111467. Genes (Basel). 2024. PMID: 39596667 Free PMC article.
-
Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency.Clin Chem. 2015 Feb;61(2):412-9. doi: 10.1373/clinchem.2014.231019. Epub 2014 Dec 11. Clin Chem. 2015. PMID: 25502182 Free PMC article.
-
High Incidence of Severe Combined Immunodeficiency Disease in Saudi Arabia Detected Through Combined T Cell Receptor Excision Circle and Next Generation Sequencing of Newborn Dried Blood Spots.Front Immunol. 2018 Apr 16;9:782. doi: 10.3389/fimmu.2018.00782. eCollection 2018. Front Immunol. 2018. PMID: 29713328 Free PMC article.
-
Newborn Screening through TREC, TREC/KREC System for Primary Immunodeficiency with limitation of TREC/KREC. Comprehensive Review.Antiinflamm Antiallergy Agents Med Chem. 2021;20(2):132-149. doi: 10.2174/1871523019999200730171600. Antiinflamm Antiallergy Agents Med Chem. 2021. PMID: 32748762 Review.
-
The case for newborn screening for severe combined immunodeficiency and related disorders.Ann N Y Acad Sci. 2011 Dec;1246:108-17. doi: 10.1111/j.1749-6632.2011.06346.x. Ann N Y Acad Sci. 2011. PMID: 22236435 Free PMC article. Review.
References
-
- Guthrie R, Susi A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 1963;32: 338–343. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

