Glycolysis in hepatic stellate cells coordinates fibrogenic extracellular vesicle release spatially to amplify liver fibrosis
- PMID: 38941469
- PMCID: PMC11212729
- DOI: 10.1126/sciadv.adn5228
Glycolysis in hepatic stellate cells coordinates fibrogenic extracellular vesicle release spatially to amplify liver fibrosis
Abstract
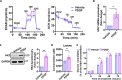
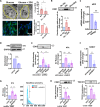
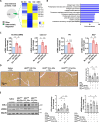
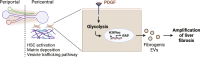
Liver fibrosis is characterized by the activation of perivascular hepatic stellate cells (HSCs), the release of fibrogenic nanosized extracellular vesicles (EVs), and increased HSC glycolysis. Nevertheless, how glycolysis in HSCs coordinates fibrosis amplification through tissue zone-specific pathways remains elusive. Here, we demonstrate that HSC-specific genetic inhibition of glycolysis reduced liver fibrosis. Moreover, spatial transcriptomics revealed a fibrosis-mediated up-regulation of EV-related pathways in the liver pericentral zone, which was abrogated by glycolysis genetic inhibition. Mechanistically, glycolysis in HSCs up-regulated the expression of EV-related genes such as Ras-related protein Rab-31 (RAB31) by enhancing histone 3 lysine 9 acetylation on the promoter region, which increased EV release. Functionally, these glycolysis-dependent EVs increased fibrotic gene expression in recipient HSC. Furthermore, EVs derived from glycolysis-deficient mice abrogated liver fibrosis amplification in contrast to glycolysis-competent mouse EVs. In summary, glycolysis in HSCs amplifies liver fibrosis by promoting fibrogenic EV release in the hepatic pericentral zone, which represents a potential therapeutic target.
Figures




References
-
- Asrani S. K., Devarbhavi H., Eaton J., Kamath P. S., Burden of liver diseases in the world. J. Hepatol. 70, 151–171 (2019). - PubMed
-
- Bhattacharya M., Ramachandran P., Immunology of human fibrosis. Nat. Immunol. 24, 1423–1433 (2023). - PubMed
-
- Ramachandran P., Matchett K. P., Dobie R., Wilson-Kanamori J. R., Henderson N. C., Single-cell technologies in hepatology: New insights into liver biology and disease pathogenesis. Nat. Rev. Gastroenterol. Hepatol. 17, 457–472 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

