Biomarker screen for efficacy of oncolytic virotherapy in patient-derived pancreatic cancer cultures
- PMID: 38941955
- PMCID: PMC11260584
- DOI: 10.1016/j.ebiom.2024.105219
Biomarker screen for efficacy of oncolytic virotherapy in patient-derived pancreatic cancer cultures
Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) is a tumour entity with unmet medical need. To assess the therapeutic potential of oncolytic virotherapy (OVT) against PDAC, different oncolytic viruses (OVs) are currently investigated in clinical trials. However, systematic comparisons of these different OVs in terms of efficacy against PDAC and biomarkers predicting therapeutic response are lacking.
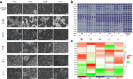
Methods: We screened fourteen patient-derived PDAC cultures which reflect the intra- and intertumoural heterogeneity of PDAC for their sensitivity to five clinically relevant OVs, namely serotype 5 adenovirus Ad5-hTERT, herpes virus T-VEC, measles vaccine strain MV-NIS, reovirus jin-3, and protoparvovirus H-1PV. Live cell analysis, quantification of viral genome/gene expression, cell viability as well as cytotoxicity assays and titration of viral progeny were conducted. Transcriptome profiling was employed to identify potential predictive biomarkers for response to OV treatment.
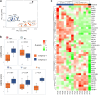
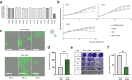
Findings: Patient-derived PDAC cultures showed individual response patterns to OV treatment. Twelve of fourteen cultures were responsive to at least one OV, with no single OV proving superior or inferior across all cultures. Known host factors for distinct viruses were retrieved as potential biomarkers. Compared to the classical molecular subtype, the quasi-mesenchymal or basal-like subtype of PDAC was found to be more sensitive to H-1PV, jin-3, and T-VEC. Generally, expression of viral entry receptors did not correlate with sensitivity to OV treatment, with one exception: Expression of Galectin-1 (LGALS1), a factor involved in H-1PV entry, positively correlated with H-1PV induced cell killing. Rather, cellular pathways controlling immunological, metabolic and proliferative signaling appeared to determine outcome. For instance, high baseline expression of interferon-stimulated genes (ISGs) correlated with relative resistance to oncolytic measles virus, whereas low cyclic GMP-AMP synthase (cGAS) expression was associated with exceptional response. Combination treatment of MV-NIS with a cGAS inhibitor improved tumour cell killing in several PDAC cultures and cells overexpressing cGAS were found to be less sensitive to MV oncolysis.
Interpretation: Considering the heterogeneity of PDAC and the complexity of biological therapies such as OVs, no single biomarker can explain the spectrum of response patterns. For selection of a particular OV, PDAC molecular subtype, ISG expression as well as activation of distinct signaling and metabolic pathways should be considered. Combination therapies can overcome resistance in specific constellations. Overall, oncolytic virotherapy is a viable treatment option for PDAC, which warrants further development. This study highlights the need for personalised treatment in OVT. By providing all primary data, this study provides a rich source and guidance for ongoing developments.
Funding: German National Science Foundation (Deutsche Forschungsgemeinschaft, DFG), German Cancer Aid (Deutsche Krebshilfe), German National Academic Scholarship Foundation (Studienstiftung des deutschen Volkes), Survival with Pancreatic Cancer Foundation.
Keywords: Cancer immunotherapy; Oncolytic virotherapy; Pancreatic cancer; Viral vectors.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests A.M. is inventor on several PV-related patents and patent applications. G.U. acts as CMO, CSO, and COO of CanVirex, a company developing oncolytic viruses for cancer therapy. C.E.E. is listed as inventor on patent applications filed by her institution related to the development of RNA viruses for cancer immunotherapy. S.H. reports an advisory role and honoraria from Amgen. L.J.A.C.H. received a grant from Flanders Innovation & Entrepreneurship (HBC18-2002) for the development of a PDXO drug screening platform.
Figures



References
-
- Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395(10242):2008–2020. - PubMed
-
- Lanson N.A., JR., Friedlander P.L., Schwarzenberger P., Kolls J.K., Wang G. Replication of an adenoviral vector controlled by the human telomerase reverse transcriptase promoter causes tumor-selective tumor lysis. Cancer Res. 2003;63(22):7936–7941. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

