The centrosomal protein FGFR1OP controls myosin function in murine intestinal epithelial cells
- PMID: 38942017
- PMCID: PMC11421975
- DOI: 10.1016/j.devcel.2024.06.001
The centrosomal protein FGFR1OP controls myosin function in murine intestinal epithelial cells
Abstract
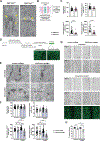
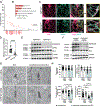
Recent advances in human genetics have shed light on the genetic factors contributing to inflammatory diseases, particularly Crohn's disease (CD), a prominent form of inflammatory bowel disease. Certain risk genes associated with CD directly influence cytokine biology and cell-specific communication networks. Current CD therapies primarily rely on anti-inflammatory drugs, which are inconsistently effective and lack strategies for promoting epithelial restoration and mucosal balance. To understand CD's underlying mechanisms, we investigated the link between CD and the FGFR1OP gene, which encodes a centrosome protein. FGFR1OP deletion in mouse intestinal epithelial cells disrupted crypt architecture, resulting in crypt loss, inflammation, and fatality. FGFR1OP insufficiency hindered epithelial resilience during colitis. FGFR1OP was crucial for preserving non-muscle myosin II activity, ensuring the integrity of the actomyosin cytoskeleton and crypt cell adhesion. This role of FGFR1OP suggests that its deficiency in genetically predisposed individuals may reduce epithelial renewal capacity, heightening susceptibility to inflammation and disease.
Keywords: FGFR1OP; adhesion; autoimmunity; centrosome; cytoskeleton; desmosome; epithelial cells; inflammatory bowel disease; non-muscle myosin.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.J.X. is co-founder of Jnana Therapeutics and Celsius Therapeutics, scientific advisory board member at Nestlé, and board director at MoonLake Immunotherapeutics. These organizations had no roles in this study.
Figures





References
-
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, and Clevers H (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. 10.1016/j.cell.2010.09.016. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

