The role of Mce proteins in Mycobacterium avium paratuberculosis infection
- PMID: 38942800
- PMCID: PMC11213854
- DOI: 10.1038/s41598-024-65592-2
The role of Mce proteins in Mycobacterium avium paratuberculosis infection
Abstract
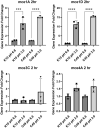
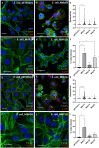
Mycobacterium avium subspecies paratuberculosis (MAP) is the causative agent of Johne's Disease, a chronic granulomatous enteritis of ruminants. MAP establishes an infection in the host via the small intestine. This requires the bacterium to adhere to, and be internalised by, cells of the intestinal tract. The effector molecules expressed by MAP for this purpose remain to be fully identified and understood. Mammalian cell entry (mce) proteins have been shown to enable other Mycobacterial species to attach to and invade host epithelial cells. Here, we have expressed Mce1A, Mce1D, Mce3C and Mce4A proteins derived from MAP on the surface of a non-invasive Escherichia coli to characterise their role in the initial interaction between MAP and the host. To this end, expression of mce1A was found to significantly increase the ability of the E. coli to attach and survive intracellularly in human monocyte-like THP-1 cells, whereas expression of mce1D was found to significantly increase attachment and invasion of E. coli to bovine epithelial cell-like MDBK cells, implying cell-type specificity. Furthermore, expression of Mce1A and Mce1D on the surface of a previously non-invasive E. coli enhanced the ability of the bacterium to infect 3D bovine basal-out enteroids. Together, our data contributes to our understanding of the effector molecules utilised by MAP in the initial interaction with the host, and may provide potential targets for therapeutic intervention.
Keywords: Mycobacterium avium ssp paratuberculosis; Enteroids; MAP; Mammalian cell entry gene; Microbial-cell interaction; Mycobacteria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Larsen AB, Merkal RS, Cutlip RC. Age of cattle as related to resistance to infection with Mycobacterium paratuberculosis. Am. J. Vet. Res. 1975;36(3):255–257. - PubMed
-
- Hase K, Kawano K, Nochi T, Pontes GS, Fukuda S, Ebisawa M, Kadokura K, Tobe T, Fujimura Y, Kawano S, Yabashi A, Waguri S, Nakato G, Kimura S, Murakami T, Iimura M, Hamura K, Fukuoka S, Lowe AW, Itoh K, Kiyono H, Ohno H. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature. 2009;462(7270):226–230. doi: 10.1038/nature08529. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

